Wei Liu
Email: wheiliu@163.com
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 5 ISSUE: 2
Page No: 66-82
Wei Liu
Email: wheiliu@163.com
Zhen Li1, Qingguo Wang1, Jiaowen Pan1, Pengfei Wang3, Guilin Zhang1, Lianqun Yang1, Fangyin Yao1, 2, Wei Liu1, 2*
1 Biotechnology Research Center, Shandong Academy of Agricultural Sciences / Key Laboratory of Genetic Improvement, Ecology and Physiology of Crops, Jinan 250100, Shandong, China
2 College of Life Sciences, Shandong Normal University, Jinan 250100, Shandong, China
3 Shandong Academy of Grape; Shandong engineering research center for Grape cultivation and deep-processing, Jinan 250100, Shandong, China
Xiujun Zhang(zhangxj@wbgcas.cn)
Xiaoxu Li(82101171073@caas.cn)
Haifeng Li(lhf@nwsuaf.edu.cn)
Yuehui Tang(yhtang2005@163.com)
Wei Liu, Genome-wide characterization of Rice Black Streaked Dwarf Virus-responsive genes in rice (2020) Journal of Food Science & Technology 5(2) pp:66-82
Background: Rice black streaked dwarf virus (RBSDV) is an important pathogen disease in rice and gramineous planting regions all over the world. The major phenotypes of RBSDV infected rice were dwarf with dark green leaves, which generally resulted in seriously loss of yield. The RBSDV known so far is preoperatively transmitted to rice in a persistent manner by small brown planthopper (SBPH), instead of transmitting to offspring through ovary.
Results: To identify RBSDV responsive genes and to explore and clarify the molecular mechanisms involved in plant-virus interaction, digital gene expression profile (DGE) analysis was performed by high-throughput sequencing using wild-type (WT) and RBSDV infected rice leaves (IRL) as materials. A total of 165,975 and 165,940 unique tags were obtained in IRL and WT libraries, respectively. In comparison with the control, 896 differentially expressed genes (DEGs) were obtained, of which 500 DEGs were up regulated and 396 DEGs were down regulated. Functional analysis showed that DEGs mainly classified into10 groups, including metabolism, stress pathogen and defense, signal transduction, Transporter, transcription and post-transcription, cell structure and division etc. To further validate reliability and authenticity of the data, 10 DEGs were randomly picked and Real-time RT-PCR was carried out, and the expression trends of 7 genes were in line with the RNA-seq results. By searching the RBSDV related miRNA database of rice, 10 targeted genes of 6 significantly changed miRNAs were also identified in these DEGs.
Conclusions: The data derived from RNA-seq were valid and credible. Through this research, a series of candidate RBSDV-responsive genes were obtained, and special signal transduction and metabolism pathways were built and pulled out in rice. This study provided further insight into the molecular mechanisms during compatible and incompatible interactions between viruses and their host plants.
Rice black streaked dwarf virus (RBSDV), which belongs to the genus Fijivirus within the family Reoviridae, could propagated and infected rice, barley, wheat, sorghum, maize and other host plants by planthopper laodelphax striatrllus in a persistent manner. The harmful influence of corresponding disease is reported to cause extensive and devastating crop losses in grain and forage yield, which ultimately leading serious economic loss in Asian countries such as China and Japan [1]. The infection of rice by RBSDV usually cause distant disease symptoms and growth abnormalities, such as severe dwarfing, more ineffective tillerings, poorly developed roots, rigidity leaf with almost no heading [2].
The whole genome of RBSDV consists of 10 double-stranded RNA segments (S1–S10) [3,4]. The complete nucleotide sequences of these segments have been determined and the functions of encoding proteins also have been preliminary researched. The mechanism by which many proteins are involved in plant virus interaction has also been studied. For instance, S7 contains two ORFs which encoding p7-1 and p7-2 proteins. Therein P7-1 may be disruption of the lignin biosynthesis and H2O2-dependent polymerization pathways, and P7-2 might involve in the plant-virus interaction [5,6]. S8 encodes core capsid protein, which may act as a nuclear transcriptional inhibitory factor in plant cells that participated in host cell gene expression regulation to make it suitable for virus propagation[1]. Although the biological functions of viral genome becoming increasingly clear, there were still no effective strategies to monitoring and control this disease as the insufficient data of the interactions between RBSDV and its host plants.
Microarray investigation revealed that various disease resistance genes, cell wall-related genes and development-related genes were altered in RBSDV infected maize[7]. Proteomic investigation also revealed that the dramatic changes of the basal metabolism processes in infected maize. Moreover, the demands for antioxidant enzymes, G-proteins, lipoxygenases and UDP-glucosyltransferase BX9 all increased in the virus infected plants, which indicated that they may participate in disease resistant response[2]. Using comparative proteomic analysis, 72 differently expressed proteins which belonged to 11 groups were detected in RBSDV permanently infected rice[8]. Using deep sequencing approach, the expression of 14 miRNAs in leave and 16 miRNAs in root were found significantly changed in RBSDV infected rice [9]. Genetic analysis also showed that the resistance of rice variety Tetep to RBSDV was controlled by quantitative trait locis (QTLs), and three new QTLs were also identified [10].
High-throughput sequencing technology has been used to researches of various virus infected plant species, such as Arabidopsis thaliana [11,12], Nicotiana [13], Zea may [14], potato[15] and rice[16]. As a model plant, the multiple genome and transcriptome databases and bioinformatical resources of rice are already available, which make it practicable to carry out high-throughput sequencing.
In this research, using WT and the RBSDV infected rice leaves (IRL) as materials, the genes expression profiles were investigated by RNA-seq, and the physiological and biochemical indexes were detected. Finally, about 896 DEGs were identified, and partial expression patterns were further verified. The expression patterns of several disease-related miRNAs targeted genes were also detected. The expression profiles or transcriptomes of the genes have provided effective clues for plants disease resistance breeding and germplasm innovation.
2.1. Plant materials
Epidemiological investigation of RBSDV was carried out in Jining, Shandong. As the production and quality of RBSDV susceptible rice variety “Zixiangnuo 2315” were heavily infected by RBSDV infection, ten RBSDV infected rice plants with symptoms were picked while the healthy strains were harvested as control locally (Figure 1). All of the rice samples were stored in -80℃ for further analysis. To verify whether the dwarf symptoms were caused by RBSDV infection, two pairs of specific PCR primers (supplement table 2) of published virus S9 and S10 were designed.

Fig1.The phenotypes of rice at tillering stage. Left: Wild type; Right: Rice infected with RBSDV
2.2. RNA extraction and cDNA libray construction
Total RNA of WT and IRL were extracted using Trizol reagent, and the crude extracts were treated with RNase-free DNaseⅠ(TaKaRa, Dalian, China). In order to reduce the individual error, the RNA of 3 independent strains of RBSDV infected or wild type plants were mixed as one sample. The total RNA concentration was examined with a spectrophotometer (Nanodrop ND-2000, ThermoFisher Scientific, Wilmington, DE, USA), and the RNA sample integrity was verified by a Bio-Analyzer 2100 (Agilent Technologies, Santa.Clara, USA). The cDNA libraries construction and Sequencing at BGI Tech Solutions Co., Ltd. (BGI Tech, Shenzhen, China).
2.3. Identification of differential expressed genes (DEGs)
Obtaining and screening the unambiguous clean tags, the number of unambiguous clean tags for each gene were calculated and then normalized to TPM (number of transcripts per million clean tags).
The DEGs between two samples were identified by a rigorous algorithm method. The "FDR (False Discovery Rate)≤0.001 and the absolute value of log2Ratio≥1" were used as the threshold to judge the significance of gene expression difference. The biological replication was analysis using Pearson correlation. According to the correlation results, DEGs were selected based on the NOISeq method of Q≥0.6 [17].
2.4. Gene Ontology (GO) and pathway enrichment analysis of DEGs
GO and pathway annotation and enrichment analysis were based on the Gene Ontology Database (http://www.geneontology.org/) and KEGG pathway Database (http://genome.jp/kegg/).
2.5. Quantitative Real-time PCR (qRT- PCR) Analysis
The gene-specific primers were designed using primer3.0 plus (supplement table 2). The qrt-PCR was performed using FastStart Universal SYBR Green Master (Roch, Shanghai, China) according to the manufacturer’s instructions. Each reaction was performed refer to Liu Wei report[18]. The rice actin gene (LOC_Os03g50885) was used as internal control. Quantification of the relative changes in gene expressions were performed using the 2-△△CT method as described by Livak and Schmittgen [19].
2.6. MicroRNA expression analysis
Refered to the data of RBSDV responsive miRNA-targeted genes [9], the putative target miRNAs of DEGs in this research were obtained.
3.1 Verification of virus infected rice strains
The RBSDV susceptible rice that sampling from Jining were preliminary screened according to Li Shuo(2015) report [20,21]. After been screened by specific primers of virus segments of S8 and S9 by RT-PCR, eight RBSDV infected rice strains with notable stunting symptoms and nine healthy strains were obtained (Supplement figure 1).
Considering RBSDV and rice stripe virus (RSV) all can be transmitted by planthopper laodelphax striatrllus simultaneously, the RSV specific primers (Supplement table 2) were also used to distinguish single virus infection or co-infection. The results showed that only one strain was detected to be infected with both RBSDV and RSV, other strains were all RBSDV single virus infection.
3.2 Tags identification and quantification
After library construction and RNA-seq analysis, total of 4,069,983 and 4,854,614 tags were obtained separately in IRL and WT. There about 3,662,063 and 4,559,710 clean tags were left after per-filter. Then, only tags with less than one copy remain in the library and were used for further analysis. Finally, 165,975 and 165,940 unique tags for the IRL and WT were obtained, respectively. All clean tags were mapped to the reference genes and genome of rice. There were 65,457 (39.45% of clean tags) unique tags in WT library and 57,703 (34.77% of clean tags) unique tags in IRL library, and the unique tags were mapped to 17,785 (26.81%) and 17424 (26.27%) of the reference genes, separately (Table 1). Meanwhile, 41,422 (24.96%) and 40,048 (24.13%) unique tags were mapped to reference genome (Table 1).
Table 1. Distribution of tags in two DEG libraries
|
Category |
Parameter |
IRL |
WT |
|
Raw Data |
Total |
4069983 |
4854614 |
|
Distinct Tag |
467951 |
416253 |
|
|
Clean Tag |
Total number |
3662063 |
4559710 |
|
Distinct Tag number |
165975 |
165940 |
|
|
All Tag Mapping to Gene |
Total number |
3014868 |
3961765 |
|
Total % of clean tag |
82.33% |
86.89% |
|
|
Distinct Tag number |
91293 |
104584 |
|
|
Distinct Tag % of clean tag |
55.00% |
63.03% |
|
|
Unambiguous Tag Mapping to Gene |
Total number |
1808480 |
2304034 |
|
Total % of clean tag |
49.38% |
50.53% |
|
|
Distinct Tag number |
57703 |
65457 |
|
|
Distinct Tag % of clean tag |
34.77% |
39.45% |
|
|
All Tag-mapped Genes |
number |
39198 |
39706 |
|
% of ref genes |
59.09% |
59.85% |
|
|
Unambiguous Tag-mapped Genes |
number |
17424 |
17785 |
|
% of ref genes |
26.27% |
26.81% |
|
|
Mapping to Genome |
Total number |
349285 |
415276 |
|
Total % of clean tag |
9.54% |
9.11% |
|
|
Distinct Tag number |
40048 |
41422 |
|
|
Distinct Tag % of clean tag |
24.13% |
24.96% |
|
|
Unknown Tag |
Total number |
297910 |
182669 |
|
Total % of clean tag |
8.14% |
4.01% |
|
|
Distinct Tag number |
34634 |
19934 |
|
|
Distinct Tag % of clean tag |
20.87% |
12.01% |
3.3 Gene expression analysis between WT and IRL libraries
About 896 DEGs were identified between WT and IRL libraries, among which 500 genes were up regulated and 396 genes were down regulated. The DEGs were mainly divided into 10 groups, among which more than 50% of DEGs were involved in stress and defense response and metabolic processes. Other DEGs were classified into hormone biosynthesis and signal transduction, Protein biosynthesis, cell structure and division (Figure 3).
Considering the screening of the disease resistance genes and breeding of disease resistance rice varieties, the categories of photosynthesis-related DEGs, hormone biosynthesis and signal transduction related DEGs, pathogenesis-related DEGs, and other remarkable DEGs were analyzed emphatically.
3.3.1 Photosynthesis-related DEGs
Here totally 23 photosynthesis-related DEGs were down- regulated in RBSDV-infected rice. Among which 18 DEGs belong to photosystem II reaction center protein, chlorophy Ⅱ A-B binding protein, photosystem I reaction center protein, photosynthetic electron transport and F-type ATPase (Table 2). There were 5 DEGs that function in the Calvin-Benson cycle, including two fructose-1,6-bisphosphatase, one phosphoribulokianse/Uridine kinase family protein, one ribulose bisphosphate carboxylase small chain, and one phosphoglycerate kinase protein (Table 2 and Table 3) .
Table 2. Photosynthesis-related genes
|
|
Locus1 |
Annotation |
Log2Ratio2 (IRL/WT) |
Q value3 |
|
Photosystem Ⅱ(PSⅡ) |
LOC_Os04g16770.1 |
photosynthetic reaction center protein, putative, expressed |
-1.81 |
0.74 |
|
LOC_Os08g25900.1 |
PsbP, putative, expressed |
-1.21 |
0.66 |
|
|
LOC_Os08g02630.1 |
photosystem II core complex proteins psbY, chloroplast precursor, putative, expressed |
-1.38 |
0.74 |
|
|
LOC_Os03g21560.1 |
photosystem II 11 kD protein, putative, expressed |
-1.82 |
0.73 |
|
|
Light harvesting complexes of PSⅡ |
LOC_Os09g17740.1 |
chlorophyll A-B binding protein, putative, expressed |
-1.44 |
0.75 |
|
LOC_Os01g52240.1 |
chlorophyll A-B binding protein, putative, expressed |
-1.43 |
0.75 |
|
|
LOC_Os07g37550.1 |
chlorophyll A-B binding protein, putative, expressed |
-1.76 |
0.70 |
|
|
LOC_Os04g38410.1 |
chlorophyll A-B binding protein, putative, expressed |
-1.66 |
0.76 |
|
|
LOC_Os09g17740.1 |
|chlorophyll A-B binding protein, putative, expressed |
-1.44 |
0.75 |
|
|
Photosystem Ⅰ(PSⅠ) |
LOC_Os08g44680.1 |
photosystem I reaction center subunit II, chloroplast precursor, putative, expressed |
-1.35 |
0.74 |
|
LOC_Os09g30340.1 |
photosystem I reaction center subunit, chloroplast precursor, putative, expressed |
-1.25 |
0.72 |
|
|
LOC_Os12g08770.1 |
photosystem I reaction center subunit N, chloroplast precursor, putative, expressed |
-1.12 |
0.70 |
|
|
LOC_Os04g33830.1 |
membrane protein, putative, expressed |
-1.80 |
0.78 |
|
|
Photosynthetic Electron transport |
LOC_Os06g01210.1 |
plastocyanin, chloroplast precursor, putative, expressed |
-1.54 |
0.77 |
|
LOC_Os06g01850.1 |
ferredoxin--NADP reductase, chloroplast precursor, putative, expressed |
-1.52 |
0.73 |
|
|
F-type ATPase |
LOC_Os07g32880.1 |
ankyrin repeat domain containing protein, expressed |
-1.28 |
0.71 |
|
LOC_Os03g17070.1 |
cysteine-rich repeat secretory protein 55 precursor, putative, expressed |
-1.23 |
0.71 |
1 based on Rice Genome Annotation Project database
2 Log2-based differential expression ratio
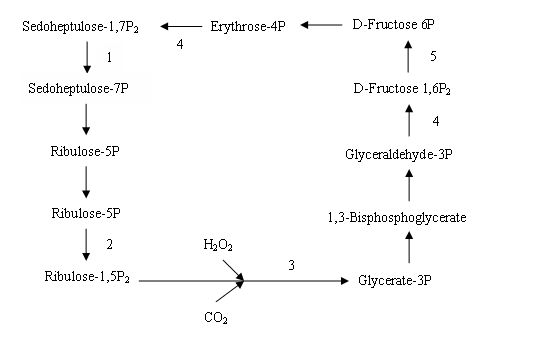
Table 3. Calvin-Benson cycle related genes
|
Designation |
Locus1 |
Annotation |
Log2Ratio2 (IRL/WT) |
Q value |
|
1 |
LOC_Os04g16680.1 |
fructose-1,6-bisphosphatase, putative, expressed |
-1.38 |
0.73 |
|
2 |
LOC_Os02g47020.1 |
phosphoribulokinase/Uridine kinase family protein, expressed |
-1.12 |
0.69 |
|
3 |
LOC_Os12g19381.1 |
ribulose bisphosphate carboxylase small chain, chloroplast precursor, putative, expressed |
-1.41 |
0.75 |
|
4 |
LOC_Os12g07210.1 |
serine esterase family protein, putative, expressed |
-1.08 |
0.65 |
|
5 |
LOC_Os03g16050.1 |
fructose-1,6-bisphosphatase, putative, expressed |
-1.88 |
0.67 |
1 based on Rice Genome Annotation Project database
2 Log2-based differential expression ratio
3.3.2 Synthesis, metabolism and signal transduction related DEGs of plant hormones
Considering the dwarf and stunted phenotypes of RBSDV infected rice, multiple clues all indicated that RBSDV disease is closely related to the synthesis and metabolism of plant hormones[22-24]. The genes involved in corresponded processes and signal transduction pathways were focused.
Genes related to Gibberellins biosynthesis and signaling
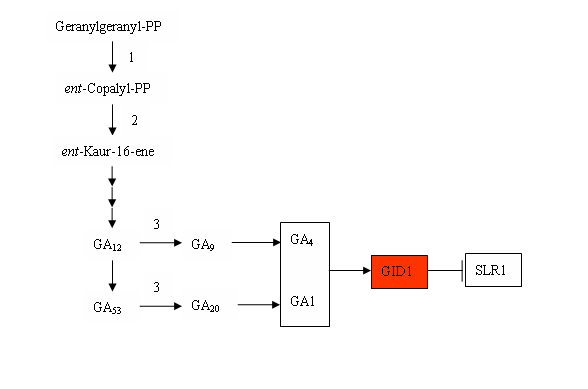
Here three significantly DEGs of GA biosynthesis were identified. The expression decreased genes mainly included one ent-copaly1 diphosphate synthase (CPS) and two gibberellins 20 oxidase 2 (GA20ox 2). Meanwhile, the expression levels of four genes encoding GID1 were increased (Table 4), which known to be involved in GA signaling pathway.
Table 4. Genes related to Gibberellins biosynthesis and signaling
|
Designation |
Locus 1 |
Annotation |
Log2Ratio2 (IRL/WT) |
Q value |
|
1 |
LOC_Os09g15050.1 |
ent-kaurene synthase A, chloroplast precursor, putative, expressed |
-2.34 |
0.69 |
|
2
|
LOC_Os04g10060.1 |
ent-kaurene synthase, chloroplast precursor, putative, expressed |
8.38 |
0.69 |
|
LOC_Os02g36140.2 |
terpene synthase, putative, expressed |
3.39 |
0.79 |
|
|
3 |
LOC_Os05g34854.1 |
gibberellin 20 oxidase 2, putative, expressed |
-1.38 |
0.73 |
|
LOC_Os05g34854.2 |
gibberellin 20 oxidase 2, putative, expressed |
-7.87 |
0.62 |
|
|
GID1
|
LOC_Os03g57640.1 |
gibberellin receptor GID1L2, putative, expressed |
1.62 |
0.74 |
|
LOC_Os03g15270.1 |
gibberellin receptor GID1L2, putative, expressed |
1.10 |
0.69 |
|
|
LOC_Os11g04350.1 |
cell death associated protein, putative, expressed |
9.56 |
0.83 |
|
|
LOC_Os07g34280.1 |
CXE carboxylesterase, putative, expressed |
2.38 |
0.78 |
1 based on Rice Genome Annotation Project database
2 Log2-based differential expression ratios
Boxes of red indicate that the corresponding genes are predominantly induced.
Genes related to brassinolide biosynthesis and signaling
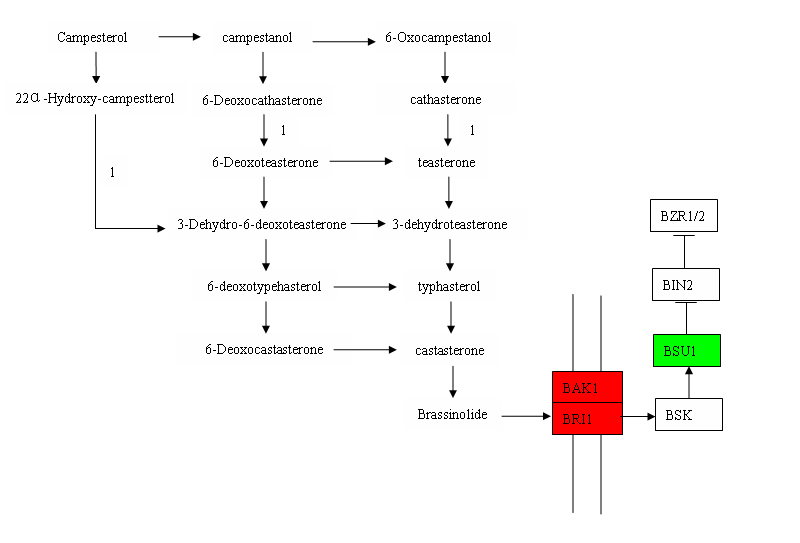
Two DEGs encoding cytochrome P450 were induced to express by RBSDV which identified to be involved in BRs biosynthesis. In the BRs signal transduction pathway, the BRASSONOSTEROID -INSENSITIVE 1 (BRI1), BRI1 ASSOCIATED RECEPTOR KINASE1 (BAK1) and BSU1 were all found to be up-regulated under disease infected conditions (Table 5).
In addition, there were two DEGs which participate in BR biosynthesis regulation. One gene named OsCYP51G3 which may encode obtusifoliol 14α-demethylase was up-regulated. Previous investigation showed that decreased OsCYP51G3 expression could reduce phytosterol and BR concentrations, and cause typical phenotypic changes related to phytosterol and BR deficiency, including dwarf plants, erect leaves, semi-sterile pollen grains, and shorter cells[25].Other gene OsGSR1 was down-regulated, which could activate BR synthesis by directly regulating a BR biosynthetic enzyme at the post-translational level and regulate GA signaling as a positive regulator [26].
Table 5. Genes related to brassinolide biosynthesis and signaling
|
Designation |
Locus1 |
Annotation |
Log2Ratio2 (IRL/WT) |
Q value |
|
1
|
LOC_Os01g43710.1 |
WRKY11, expressed |
1.14 |
0.70 |
|
LOC_Os04g48200.1 |
cytochrome P450, putative, expressed |
8.28 |
0.67 |
|
|
BRI1
|
LOC_Os11g39420.1 |
jacalin-like lectin domain containing protein, expressed |
4.12 |
0.73 |
|
LOC_Os04g42620.1 |
uncharacterized protein At4g06744 precursor, putative, expressed |
-1.03 |
0.65 |
|
|
BAK1
|
LOC_Os01g04580.1 |
Ser/Thr protein kinase, putative, expressed |
1.60 |
0.66 |
|
LOC_Os02g48080.1 |
cysteine-rich receptor-like protein kinase 7 precursor, putative, expressed |
1.18 |
0.60 |
|
|
LOC_Os01g12430.1 |
receptor-like protein kinase, putative, expressed |
1.40 |
0.60 |
|
|
LOC_Os01g12420.1 |
expressed protein |
1.92 |
0.61 |
|
|
LOC_Os07g43570.1 |
TKL_IRAK_DUF26-lc.25 - DUF26 kinases have homology to DUF26 containing loci, expressed |
-1.31 |
0.68 |
|
|
LOC_Os10g30530.1 |
lectin-like receptor kinase, putative, expressed |
1.39 |
0.60 |
|
|
BSU1 |
LOC_Os09g07460.1 |
fasciclin-like arabinogalactan protein 8 precursor, putative, expressed |
1.09 |
0.61 |
1 based on Rice Genome Annotation Project database
2 Log2-based differential expression ratios
Boxes of red indicate that the corresponding genes are predominantly induced.
Boxes of green indicate that the corresponding genes are predominantly suppressed.
3.3.3 Pathogenesis-related DEGs.
The results showed that 75 pathogenesis-related DEGs were identified, among them 61 genes up-regulated and 14 genes were down-regulated after infection. Specifically, there were 7 SCP-like extracellular protein encoded genes, 8 chitinase genes, 6 thaumatin genes, 12 glycosyl hydrolase genes, 11 peroxidase genes, 10 proteinase inhibitor genes, 7 Cupin domain containing protein genes and 14 other PR-like genes (Table 6).
Table 6. Disease resistance related genes
|
PR family |
Locus1 |
Annotation |
Log2ratio2 (IRL/WT) |
Q value |
|
PR1 |
LOC_Os07g03580.1 |
SCP-like extracellular protein, expressed |
6.64 |
0.93 |
|
|
LOC_Os07g03319.1 |
SCP-like extracellular protein, expressed |
6.27 |
0.87 |
|
|
LOC_Os10g11500.1 |
SCP-like extracellular protein, expressed |
5.40 |
0.82 |
|
|
LOC_Os07g03600.1 |
SCP-like extracellular protein, expressed |
4.99 |
0.70 |
|
|
LOC_Os07g03710.1 |
SCP-like extracellular protein, expressed |
4.06 |
0.83 |
|
|
LOC_Os07g03590.1 |
SCP-like extracellular protein, expressed |
3.84 |
0.78 |
|
|
LOC_Os01g28450.1 |
SCP-like extracellular protein, expressed |
3.14 |
0.83 |
|
PR2 |
LOC_Os01g71340.1 |
glycosyl hydrolases family 17, putative, expressed |
2.67 |
0.82 |
|
|
LOC_Os01g71820.1 |
glycosyl hydrolases family 17, putative, expressed |
2.15 |
0.78 |
|
|
LOC_Os01g71670.1 |
glycosyl hydrolases family 17, putative, expressed |
1.52 |
0.77 |
|
PR3 |
LOC_Os05g33130.1 |
CHIT17 - Chitinase family protein precursor, expressed |
2.56 |
0.82 |
|
|
LOC_Os05g33140.1 |
CHIT17 - Chitinase family protein precursor, expressed |
1.02 |
0.67 |
|
|
LOC_Os03g30470.1 |
CHIT4 - Chitinase family protein precursor, expressed |
2.64 |
0.81 |
|
|
LOC_Os03g31690.1 |
CHIT4 - Chitinase family protein precursor, expressed |
-1.14 |
0.62 |
|
|
LOC_Os06g51050.1 |
CHIT7 - Chitinase family protein precursor, expressed |
2.67 |
0.82 |
|
|
LOC_Os06g51060.1 |
CHIT7 - Chitinase family protein precursor, expressed |
1.16 |
0.71 |
|
PR4 |
LOC_Os11g37950.1 |
WIP3 - Wound-induced protein precursor, expressed |
1.39 |
0.75 |
|
|
LOC_Os11g37960.1 |
WIP4 - Wound-induced protein precursor, expressed |
3.08 |
0.831 |
|
|
LOC_Os11g37970.1 |
WIP5 - Wound-induced protein precursor, expressed |
1.52 |
0.68 |
|
PR5 |
LOC_Os12g38120.1 |
thaumatin family domain containing protein, expressed |
2.00 |
0.77 |
|
|
LOC_Os12g43450.1 |
thaumatin family domain containing protein, expressed |
1.47 |
0.76 |
|
|
LOC_Os03g45960.1 |
thaumatin, putative, expressed |
3.07 |
0.81 |
|
|
LOC_Os12g43490.1 |
thaumatin, putative, expressed |
3.04 |
0.83 |
|
|
LOC_Os12g43510.1 |
thaumatin, putative, expressed |
1.49 |
0.61 |
|
|
LOC_Os03g46070.1 |
thaumatin, putative, expressed |
1.33 |
0.74 |
|
|
LOC_Os01g62260.1 |
thaumatin, putative, expressed |
-1.04 |
0.68 |
|
PR6 |
LOC_Os01g03340.1 |
BBTI4 - Bowman-Birk type bran trypsin inhibitor precursor, expressed |
1.41 |
0.75 |
|
|
LOC_Os01g03360.1 |
BBTI5 - Bowman-Birk type bran trypsin inhibitor precursor, expressed |
2.55 |
0.82 |
|
|
LOC_Os01g03390.1 |
BBTI7 - Bowman-Birk type bran trypsin inhibitor precursor, expressed |
1.21 |
0.72 |
|
PR8 |
LOC_Os11g18730.1 |
glycosyl hydrolase family 3 protein, putative, expressed |
-1.67 |
0.76 |
|
|
LOC_Os11g47530.1 |
glycosyl hydrolase, putative, expressed |
8.3 |
0.68 |
|
|
LOC_Os11g47560.1 |
glycosyl hydrolase, putative, expressed |
3.88 |
0.69 |
|
|
LOC_Os05g15880.1 |
glycosyl hydrolase, putative, expressed |
3.76 |
0.79 |
|
|
LOC_Os11g47550.1 |
glycosyl hydrolase, putative, expressed |
3.70 |
0.61 |
|
|
LOC_Os11g47600.1 |
glycosyl hydrolase, putative, expressed |
2.35 |
0.78 |
|
|
LOC_Os11g47510.1 |
glycosyl hydrolase, putative, expressed |
1.94 |
0.69 |
|
|
LOC_Os10g28120.1 |
glycosyl hydrolase, putative, expressed |
1.94 |
0.80 |
|
|
LOC_Os11g47570.1 |
glycosyl hydrolase, putative, expressed |
1.85 |
0.62 |
|
|
LOC_Os11g47580.1 |
glycosyl hydrolase, putative, expressed |
1.79 |
0.68 |
|
|
LOC_Os01g47070.1 |
glycosyl hydrolase, putative, expressed |
1.61 |
0.77 |
|
|
LOC_Os10g28080.1 |
glycosyl hydrolase, putative, expressed |
1.43 |
0.75 |
|
|
LOC_Os05g15770.1 |
glycosyl hydrolase, putative, expressed |
1.37 |
0.74 |
|
PR9 |
LOC_Os05g04490.1 |
peroxidase precursor, putative, expressed |
2.62 |
0.76 |
|
|
LOC_Os03g13180.1 |
peroxidase precursor, putative, expressed |
1.98 |
0.71 |
|
|
LOC_Os01g22352.1 |
peroxidase precursor, putative, expressed |
1.83 |
0.77 |
|
|
LOC_Os05g06970.1 |
peroxidase precursor, putative, expressed |
1.80 |
0.72 |
|
|
LOC_Os08g01120.1 |
peroxidase precursor, putative, expressed |
1.63 |
0.63 |
|
|
LOC_Os04g59150.1 |
peroxidase precursor, putative, expressed |
1.05 |
0.62 |
|
|
LOC_Os09g29490.1 |
peroxidase precursor, putative, expressed |
-1.41 |
0.60 |
|
|
LOC_Os07g48010.1 |
peroxidase precursor, putative, expressed |
-1.74 |
0.73 |
|
|
LOC_Os01g10850.1 |
peroxidase precursor, putative, expressed |
-1.86 |
0.67 |
|
PR10 |
LOC_Os08g28670.1 |
pathogenesis-related Bet v I family protein, putative, expressed |
2.31 |
0.71 |
|
|
LOC_Os12g36850.1 |
pathogenesis-related Bet v I family protein, putative, expressed |
1.83 |
0.75 |
|
|
LOC_Os12g36830.1 |
pathogenesis-related Bet v I family protein, putative, expressed |
1.75 |
0.75 |
|
|
LOC_Os01g14590.1 |
pathogen-related protein, putative, expressed |
1.41 |
0.70 |
|
PR13 |
LOC_Os12g26960.1 |
THION34 - Plant thionin family protein precursor, expressed |
2.39 |
0.72 |
|
PR14 |
LOC_Os05g06780.1 |
LTPL104 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
-1.54 |
0.75 |
|
|
LOC_Os03g14654.1 |
LTPL108 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
1.21 |
0.69 |
|
|
LOC_Os02g44320.1 |
LTPL113 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
-1.38 |
0.74 |
|
|
LOC_Os04g55159.1 |
LTPL125 - Protease inhibitor/seed storage/LTP family protein precursor, putative, expressed |
-1.30 |
0.73 |
|
|
LOC_Os10g40420.1 |
LTPL138 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
-1.50 |
0.72 |
|
|
LOC_Os06g49190.1 |
LTPL154 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
1.61 |
0.72 |
|
|
LOC_Os10g36100.1 |
LTPL157 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
3.61 |
0.83 |
|
|
LOC_Os01g59900.1 |
LTPL65 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
-1.06 |
0.67 |
|
|
LOC_Os07g07790.1 |
LTPL75 - Protease inhibitor/seed storage/LTP family protein precursor, expressed |
2.30 |
0.66 |
|
PR15 |
LOC_Os08g35760.1 |
Cupin domain containing protein, expressed |
-1.99 |
0.80 |
|
PR16 |
LOC_Os08g08960.1 |
Cupin domain containing protein, expressed |
4.04 |
0.73 |
|
|
LOC_Os08g09000.1 |
Cupin domain containing protein, expressed |
3.83 |
0.76 |
|
|
LOC_Os08g08980.1 |
Cupin domain containing protein, expressed |
2.17 |
0.75 |
|
|
LOC_Os08g09010.1 |
cupin domain containing protein, expressed |
1.76 |
0.75 |
|
|
LOC_Os08g09060.1 |
Cupin domain containing protein, expressed |
1.08 |
0.68 |
1 based on Rice Genome Annotation Project database
2 Log2-based differential expression ratio
3.3.4 Other remarkable DEGs
Except the DGEs that classified into specific processes above, there still have 23 DEGs that present significant high expression levels, and 10 DEGs that showed decreased expression levels in RBSDV infected rice (log2Ratio IRL/WT>5) ( Supplement Table 1). Among which, there were 17 up-regulated genes and 7 down-regulated genes that have annotations in rice.
In the data, eight DGEs were defense-and stress-related genes, including MYB transcription factor, OsSub13 [27] putative subtilisin, terpene synthase [28] and DUF26 [29,30]. Two DGEs related to hormone metabolic, including GDSL-like lipase/acylhydrolase regulated of enthylene signal and flavin monooxygenase involved in auxin biosynthesis[31,32].
OsCML26, which reported to play important roles in the signal transduction and transcriptional regulation networks of Ca2+ signaling in response to different abiotic stress, was induced to express significantly (log2Ratio=8.64) after RBSDV infected [33,34].
In the data, we also found two cell wall-related genes, one was POEI5-Pollen Ole e I allergen and extensin family protein precursor (LOC_Os10g05820.1), which is a key enzyme in the biosynthesis of lignin cell wall precursors, the expression level of this gene was down-regulated after RBSDV infected (log2Ratio=-8.03). The other was pectinesterase, which is a cell-wall-associated enzyme that facilitates plant cell wall modification and subsequent breakdown. The expression level of this gene was up-regulated in infected rice (log2Ratio=7.80).
3.4. Gene Ontology (GO) enrichment analysis of DEGs
In order to understand the functions of these DEGs, all of these genes were mapped to GO database and classified into different groups according to GO terms comparing to the genome background.
In the category of molecular function (MF) the GO terms of oxidoreductase activity and binding which including pattern binding, carbohydrate binding and ion binding were significantly overrepresented, other terms such as chitinase activity were also dominant.
When looking into the category of biological process (BP), the terms of response to stress and response to biotic stimulus were found overrepresented in the IRL library, which were also found in other plants infected with pathogen. Other terms such as aminoglycan catabolic process, amine metabolic process and polysaccharide catabolic process were also dominant.
Meanwhile, in the category of cellur component (CC), an increased percentage of DEGs corresponding to the terms of plastid, thylakoid and cytoplasmic vesicle were mainly appeared in the RBSDV infected rice. The results above hinted that the regulation of photosynthesis efficiency and vesicle transportion may play important role during the plant-pathogen interaction process (Figure2).
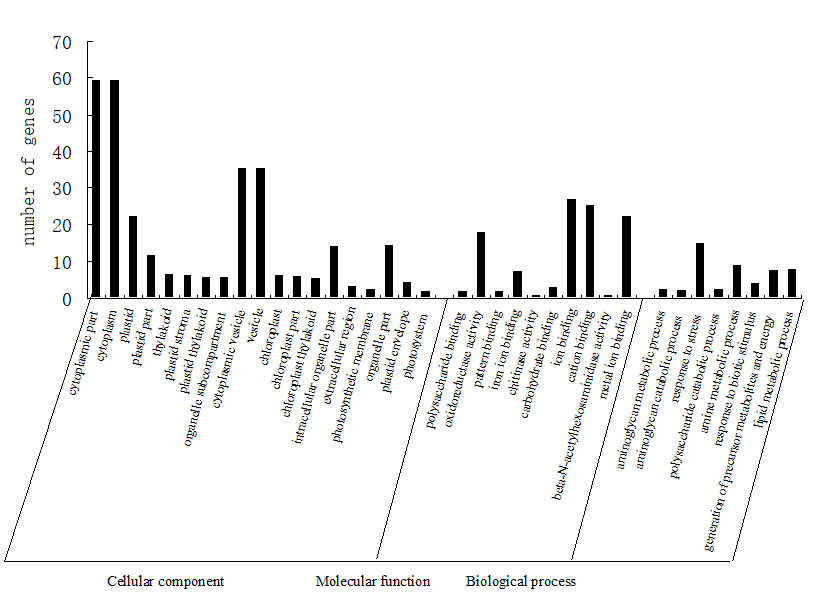
Figure 2. GO analysis of DEGs between the WT and IRL libraries.
3.5. Pathways enrichment analysis of DEGs
The DEGs identified in RBSDV infected rice were also grouped into different biological pathways by enrichment analysis. Among 896 DEGs, total of 508 DEGs were annotated in 104 pathways. Therein 19 pathways were significantly enriched (Q value≤0.05). The enriched pathways were all in metabolism and biosynthetic pathways. In metabolism pathways included carbohydrate metabolism, energy metabolism, nucleotide metabolism, lipid metabolism, metabolism of terpenoids, polyketides and biosynthesis of other secondary metabolites. The biosynthetic pathways mainly include biosynthesis of plant hormones. There were 60 DEGs belong to the biosynthesis of plant hormones, of which 44 DEGs were up-regulated and 16 DEGs were down-regulated. The biosynthesis of plant hormones pathways involved the biosynthesis of IAA, ABA, GA, JA and SA. The results suggested that RBSDV infect may cause the change of the host hormone levels, thus affect the growth and development of the host and stimulate the host disease resistance response (Figure3).
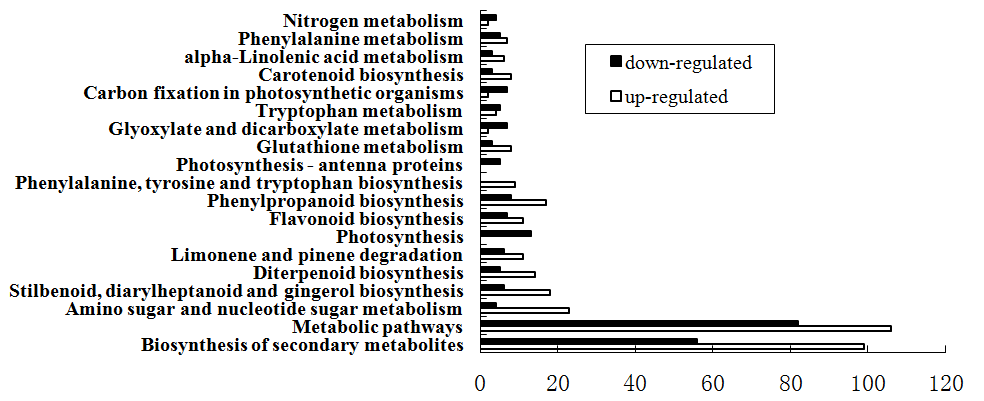
Figure 3. Pathways enrichment analysis of DEGs between the WT and IRL libraries.
3.6. Verification of profiles of DEGs
To validate the expression profiles of DEGs, 10 DEGs were picked randomly and analyzed by qRT-PCR using rice actin as internal control. The results showed that except 3 genes, the variation trends of other 7 genes were consistent with RNA-seq results (Supplement figure 2).
3.7. MicroRNA expression analysis
There were 10 DEGs targeted by miR164, miR166, miR396, miR528, miR827, miRn11. Including NAC gene, ozone-responsive apoplastic protein, OsSPX-MFS1, and so on (Table 7 ). But the expression of DEGs was notable unless ozone-responsive apoplastic protein. And the expression levels of these miRNAs will be further analyzed by qRT-PCR.
Table 7. The target DEGs of miRNA
|
miRNA |
Target gene |
Log2Ratio(IRL/WT) |
Description |
|
miR528 |
Os09g20090 |
2.85 |
ozone-responsive apoplastic protein |
|
Os02g48080 |
1.18 |
cysteine-rich receptor-like protein kinase 7 precursor, putative, express |
|
|
Os01g66170 |
-1.35 |
SNARE associated Golgi protein, putative, expressed |
|
|
miR164 |
Os12g05260 |
1.56 |
phytosulfokines precursor, putative, expressed |
|
Os06g46270 |
1.19 |
NAC gene |
|
|
Os12g41680 |
0.92 |
No apical meristem protein, putative, expressed |
|
|
miR396 |
Os02g48080 |
1.18 |
cysteine-rich receptor-like protein kinase 7 precursor, putative, express |
|
miR827 |
Os04g48390 |
1.70 |
OsSPX-MFS1 |
|
miRn11 |
Os10g39520 |
-1.12 |
MLO domain containing protein, putative, expressed |
|
miR166 |
Os04g48290 |
-0.95 |
MATE efflux family protein, putative, expressed |
4.1. Effects of RBSDV on photosynthetic rate and energy metabolism
Plant infected with RBSDV usually induces photosynthetic rate decrease. The long-term RBSDV accumulation in rice lead to the decrease of photosynthetic parameters, including the net photosynthetic rate(Pn), the stomatal conductance (Cond), the intercellular CO2 concentration (Ci), and the transpiration speed (Tr). Some related proteins such as chlorophyII a/b binding proteins, Rubisco activase small isoform precursor and calvin cycle related proteins were also changed [8]. Here, 24 DEGs that related to photosynthesis of photosystem II (PSII), photosystem I (PSI), photosynthethic electron transport, F-type ATPase and the Calvin cycle were down-regulated under virus infection.
Down regulated DEGs of photosynthetic reaction including psbA(LOC_Os04g16770.1),psbP(LOC_Os08g02630.1), psb27(LOC_Os03g21560.1), psbY(LOC_Os08g02630.1) and psbW(LOC_Os03g21560.1). The psbA encode photosynthetic reaction center protein D1, and the psbP encode subunit of the oxygen evolving complex (OEC). The decrease of the protein products of these two genes will affect the assembly and/or stability of the PSII-LHCII supercomplexes in the gram regions of the thylakoid membrane [35-37]. The PsbY protein was reported to take part in the cyclic electron flow around PSII in order to protect PSII against photoinhibition, and also could stabilize the binding of PsbE and PsbF to the heme group by providing a shelter of Cyt b559 from oxidizing compounds[38]. The psb27 was known to be essential for efficient recovery of the photodamaged PSII. In Arabidopsis, under high-light-illumination, PSII activity and the content of the PSII reaction center protein D1 decreased more significantly in the psb27 mutant than in wild-type (WT) plant[39,40]. Taken together; the decrease of these photosynthetic genes will affect the stable of PSII complex and the recovery of the photodamaged PSII.
Five Chlorophyll A-B binding protein genes were down-regulated in RBSDV infected rice. Considering chlorophyll A-B binding protein is a component of the light-harvesting complex of photosynthesis II (LHC II), down regulation of these genes may inhibit LHC II function and further affect the photosynthetic process.
In the photosystem I, there were 4 DEGs including psa D (LOC_Os08g44680.1), psaG (LOC_Os09g30340.1), psaN (LOC_Os12g08770.1) and psaO (LOC_Os04g33830.1). In view of the functions of proteins encoded by these genes, the down regulation of these genes may affect stable and activity of PSI, and influence the energy balance between photosystems [41-43]. The reduction of plastocyanin (PC) and ferredoxin--NADP reductase(FNR) may inhibit the electron transport from PS II to PSI. RBSDV infection also inhibited the carbon fixation of plants by decreasing the expressions of 5 DEGs which encoding Calvin cycle enzyme.
Taken together, the PSI, PSII and electron transport were infected in BSDV infected rice, and thereby infected the photosynthesis efficiency and energy accumulation, which further results in abnormal growth, dwarfing and fruitless symptoms of infected plants.
4.2. The relationship between dwarfing symptoms and hormones
Dwarfism is a typical characteristic symptom of RBSDV infected plants. Considering phytohormones play multiple roles in regulation growth and development of plants, their variations in RBSDV infected rice were analyzed in depth.
Gibberellins (GA) are a group of tetracyclic diterpenoid compounds, which essential for many processes of plant growth and development [44-46]. The loss of function in GA synthesis and signaling resulted in dwarfism in rice and Arabidopsis plants, and transgenic plant expressing genes for GA degradation showed the dwarfism [47-49]. In RBSDV infected rice the GA content reduces significantly [50]. In this research, partial DEGs in GA biosynthesis were decreased and in GA signaling pathway were increased, suggested that RBSDV infection might inhibite the accumulation of gibberellins, but activate GA signaling transduction in rice.
Brassinosteroid (BR) is other important phytohormone in plants. Brassinosteroids involve in various physiological processes including plant growth, development and immunity [51].In rice , a low concentration of BL (10-9M) could slightly enhance leaf sheath and root length, but it will inhibit leaf sheaths and roots growth when the contents were accumulated over certain levels, which resulted in decreased seeding height and root length [52]. Consistent with this, mutant or transgenic plants with excessive enhanced BR levels or BR signaling usually have reduced height to a certain degree in rice. In our data, two DEGs in BR biosynthesis were up-regulated significantly under virus infection, and this give us a hint that it may be caused by over accumulation of BR that lead to the dwarfism of RBSDV infected rice. The detailed mechanisms were still under investigation.
4.3. The roles of PRs in RBSDV infected rice
By now, many theoretical researches on the effects of plant PR genes have mainly focused on plant-fungi/bacteria interactions [53]. Some research found that the induced patterns of the PRs were quite different in virus infected rice, hinting the divergent defense mechanisms during defense response. In this research, 75 PRs that 61 up-regulated and 14 down-regulated were identified in the highly RBSDV susceptible rice Zixiangnuo2315. The PR have been classified into 17 groups (PR1-PR17) based on their amino acid sequences and biological functions [54].In our data, These 75 PRs belong to eleven PR subfamilies, and the DEGs of PR1, PR14 and PR16 changed significantly in this research [54]. Especially seven DEGs of PR1 (including OsPR1a and OsPR1b) were dramatically induced to express about 10 to 3000 folds, which displayed vital roles in RBSDV and host interaction[55,56]. Here ten PR genes belong to PR14 subfamily, which further defined as LTP genes were also changed, among which 6 genes were down-regulated about 3 to 285 folds and 4 genes were up-regulated about 3 to 20 folds[57]. As reported that LTP genes such as OsC6 may participate in development regulation of pollen, the phenotypes of no heading or sterility of RBSDV infected rice may association with down-regulation of LTPs[58].
This work was supported by Shandong Provential Natural Science Foundation (No. ZR2019QC003), the National Key Research and Development Program of China (2016YFD0100903-9), Agricultural scientific and technological innovation project of Shandong Academy of Agricultural Sciences (CXGC2018E13 and CXGC2016A02), and the Young Talents Training program of Shandong Academy of Agricultural Sciences (2016-2018).
Liu H, Wei C, Zhong Y, Li Y (2007) Rice black-streaked dwarf virus minor core protein P8 is a nuclear dimeric protein and represses transcription in tobacco protoplasts. FEBS Letters 581: 2534-2540. PMid:17499245
View Article PubMed/NCBILi K, Xu C, Zhang J (2011) Proteome profile of maize (Zea Mays L.) leaf tissue at the flowering stage after long-term adjustment to rice black-streaked dwarf virus infection. Gene 485: 106-113. PMid:21708230
View Article PubMed/NCBIZhang HM, Chen JP, Adams MJ (2001) Molecular characterisation of segments 1 to 6 of Rice black-streaked dwarf virus from China provides the complete genome. Archives of Virology 146: 2331-2339. PMid:11811683
View Article PubMed/NCBIWang ZH, Fang SG, Xu JL, Sun LY, Li DW, et al. (2003) Sequence analysis of the complete genome of rice black-streaked dwarf virus isolated from maize with rough dwarf disease. Virus Genes 27: 163-168. PMid:14501194
View Article PubMed/NCBISun F, Yuan X, Xu Q, Zhou T, Fan Y, et al. (2013) Overexpression of rice black-streaked dwarf virus p7-1 in Arabidopsis results in male sterility due to non-dehiscent anthers. PLoS One 8: e79514. PMid:24260239
View Article PubMed/NCBIWang Q, Tao T, Han Y, Chen X, Fan Z, et al. (2013) Nonstructural protein P7-2 encoded by Rice black-streaked dwarf virus interacts with SKP1, a core subunit of SCF ubiquitin ligase. Virol J 10: 325. PMid:24176102
View Article PubMed/NCBIJia MA, Li Y, Lei L, Di D, Miao H, et al. (2012) Alteration of gene expression profile in maize infected with a double-stranded RNA fijivirus associated with symptom development. Mol Plant Pathol 13: 251-262. PMid:21955602
View Article PubMed/NCBIXu Q, Ni H, Chen Q, Sun F, Zhou T, et al. (2013) Comparative proteomic analysis reveals the cross-talk between the responses induced by H2O2 and by long-term rice black-streaked dwarf virus infection in rice. PLoS One 8: e81640. PMid:24312331
View Article PubMed/NCBISun Z, He Y, Li J, Wang X, Chen J (2015) Genome-wide characterization of rice black streaked dwarf virus-responsive microRNAs in rice leaves and roots by small RNA and degradome sequencing. Plant Cell Physiol 56: 688-699. PMid:25535197
View Article PubMed/NCBIZhou T, Du L, Wang L, Wang Y, Gao C, et al. (2015) Genetic analysis and molecular mapping of QTLs for resistance to rice black-streaked dwarf disease in rice. Sci Rep 5: 10509. PMid:26198760
View Article PubMed/NCBIGao R, Liu P, Yong Y, Wong SM (2016) Genome-wide transcriptomic analysis reveals correlation between higher WRKY61 expression and reduced symptom severity in Turnip crinkle virus infected Arabidopsis thaliana. Sci Rep 6: 24604. PMid:27086702
View Article PubMed/NCBISun F, Fang P, Li J, Du L, Lan Y, et al. (2016) RNA-seq-based digital gene expression analysis reveals modification of host defense responses by rice stripe virus during disease symptom development in Arabidopsis. Virol J 13: 202. PMid:27912765
View Article PubMed/NCBIPesti R, Kontra L, Paul K, Vass I, Csorba T, et al. (2019) Differential gene expression and physiological changes during acute or persistent plant virus interactions may contribute to viral symptom differences. PLoS One 14: e0216618. PMid:31051010
View Article PubMed/NCBIGhorbani A, Izadpanah K, Dietzgen RG (2018) Changes in maize transcriptome in response to maize Iranian mosaic virus infection. 13: e0194592. PMid:29634778
View Article PubMed/NCBIGoyer A, Hamlin L, Crosslin JM, Buchanan A, Chang JH (2015) RNA-Seq analysis of resistant and susceptible potato varieties during the early stages of potato virus Y infection. BMC Genomics 16: 472. PMid:26091899
View Article PubMed/NCBIZheng W, Ma L, Zhao J, Li Z, Sun F, et al. (2013) Comparative transcriptome analysis of two rice varieties in response to rice stripe virus and small brown planthoppers during early interaction. PLoS One 8: e82126. PMid:24358146
View Article PubMed/NCBITarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A (2011) Differential expression in RNA-seq: a matter of depth. Genome Res 21: 2213-2223. PMid:21903743
View Article PubMed/NCBILiu W, Wang PF, Li Z, Wang QG, Wang YY, et al. (2019) Genome-Wide Identification and Expression Analysis of the Cytidine Deaminase Subfamily in Rice. Russian Journal of Plant Physiology 66: 203-213.
View ArticleLivak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402-408. PMid:11846609
View Article PubMed/NCBILi S, Wang X, Xu J, Ji Y, Zhou Y (2015) A simplified method for simultaneous detection of Rice stripe virus and Rice black-streaked dwarf virus in insect vector. J Virol Methods 211: 32-35. PMid:25455902
View Article PubMed/NCBICho SY, Jeong RD, Yoon YN, Lee SH, Shin DB, et al. (2013) One-step multiplex reverse transcription-polymerase chain reaction for the simultaneous detection of three rice viruses. J Virol Methods 193: 674-678. PMid:23850700
View Article PubMed/NCBIZhang H, Tan X, Li L, He Y, Hong G, et al. (2019) Suppression of auxin signalling promotes rice susceptibility to Rice black streaked dwarf virus infection. Mol Plant Pathol 20: 1093-1104. PMid:31250531
View Article PubMed/NCBIXie K, Li L, Zhang H, Wang R, Tan X, et al. (2018) Abscisic acid negatively modulates plant defence against rice black-streaked dwarf virus infection by suppressing the jasmonate pathway and regulating reactive oxygen species levels in rice. Plant, Cell & Environment 41: 2504-2514. PMid:29920686
View Article PubMed/NCBITao T, Zhou CJ, Wang Q, Chen XR, Sun Q, et al. (2017) Rice black streaked dwarf virus P7-2 forms a SCF complex through binding to Oryza sativa SKP1-like proteins, and interacts with GID2 involved in the gibberellin pathway. PLoS One 12: e0177518. PMid:28494021
View Article PubMed/NCBIXia K, Ou X, Tang H, Wang R, Wu P, et al. (2015) Rice microRNA osa-miR1848 targets the obtusifoliol 14alpha-demethylase gene OsCYP51G3 and mediates the biosynthesis of phytosterols and brassinosteroids during development and in response to stress. New Phytol 208: 790-802. PMid:26083975
View Article PubMed/NCBIWang L, Wang Z, Xu Y, Joo SH, Kim SK, et al. (2009) OsGSR1 is involved in crosstalk between gibberellins and brassinosteroids in rice. Plant J 57: 498-510. PMid:18980660
View Article PubMed/NCBIJordá L, Vera P (2000) Local and Systemic Induction of Two Defense-Related Subtilisin-Like Protease Promoters in Transgenic Arabidopsis Plants. Luciferin Induction of PR Gene Expression. Plant Physiology 124: 1049-1058. PMid:11080282
View Article PubMed/NCBIYoshitomi K, Taniguchi S, Tanaka K, Uji Y, Akimitsu K, et al. (2016) Rice terpene synthase 24 (OsTPS24) encodes a jasmonate-responsive monoterpene synthase that produces an antibacterial gamma-terpinene against rice pathogen. J Plant Physiol 191: 120-126. PMid:26771167
View Article PubMed/NCBIAmbawat S, Sharma P, Yadav NR, Yadav RC (2013) MYB transcription factor genes as regulators for plant responses: an overview. Physiol Mol Biol Plants 19: 307-321. PMid:24431500
View Article PubMed/NCBIMiyakawa T, Hatano K, Miyauchi Y, Suwa Y, Sawano Y, et al. (2014) A secreted protein with plant-specific cysteine-rich motif functions as a mannose-binding lectin that exhibits antifungal activity. Plant Physiol 166: 766-778. PMid:25139159
View Article PubMed/NCBIYamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T (2007) Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol 143: 1362-1371. PMid:17220367
View Article PubMed/NCBIKim HG, Kwon SJ, Jang YJ, Nam MH, Chung JH, et al. (2013) GDSL LIPASE1 modulates plant immunity through feedback regulation of ethylene signaling. Plant Physiol 163: 1776-1791. PMid:24170202
View Article PubMed/NCBIChinpongpanich A, Limruengroj K, Phean OPS, Limpaseni T, Buaboocha T (2012) Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res Notes 5: 625. PMid:23134977
View Article PubMed/NCBIBoonburapong B, Buaboocha T (2007) Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol 7: 4. PMid:17263873
View Article PubMed/NCBIThornton LE, Ohkawa H, Roose JL, Kashino Y, Keren N, et al. (2004) Homologs of plant PsbP and PsbQ proteins are necessary for regulation of photosystem ii activity in the cyanobacterium Synechocystis 6803. Plant Cell 16: 2164-2175. PMid:15258264
View Article PubMed/NCBIIdo K, Ifuku K, Yamamoto Y, Ishihara S, Murakami A, et al. (2009) Knockdown of the PsbP protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochim Biophys Acta 1787: 873-881. PMid:19285950
View Article PubMed/NCBIPlochinger M, Schwenkert S, von Sydow L, Schroder WP, Meurer J (2016) Functional Update of the Auxiliary Proteins PsbW, PsbY, HCF136, PsbN, TerC and ALB3 in Maintenance and Assembly of PSII. Front Plant Sci 7: 423. PMid:27092151
View Article PubMed/NCBIvon Sydow L, Schwenkert S, Meurer J, Funk C, Mamedov F, et al. (2016) The PsbY protein of Arabidopsis Photosystem II is important for the redox control of cytochrome b559. Biochim Biophys Acta 1857: 1524-1533. PMid:27220875
View Article PubMed/NCBIChen H, Zhang D, Guo J, Wu H, Jin M, et al. (2006) A Psb27 homologue in Arabidopsis thaliana is required for efficient repair of photodamaged photosystem II. Plant Mol Biol 61: 567-575. PMid:16897475
View Article PubMed/NCBINowaczyk MM, Hebeler R, Schlodder E, Meyer HE, Warscheid B, et al. (2006) Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of photosystem II. Plant Cell 18: 3121-3131. PMid:17114356
View Article PubMed/NCBIJensen PE, Haldrup A, Zhang S, Scheller HV (2004) The PSI-O subunit of plant photosystem I is involved in balancing the excitation pressure between the two photosystems. J Biol Chem 279: 24212-24217. PMid:15169790
View Article PubMed/NCBIChitnis VP, Ke A, Chitnis PR (1997) The PsaD subunit of photosystem I. Mutations in the basic domain reduce the level of PsaD in the membranes. Plant Physiol 115: 1699-1705. PMid:9414569
View Article PubMed/NCBIZygadlo A, Jensen PE, Leister D, Scheller HV (2005) Photosystem I lacking the PSI-G subunit has a higher affinity for plastocyanin and is sensitive to photodamage. Biochim Biophys Acta 1708: 154-163. PMid:15953472
View Article PubMed/NCBIWang Y, Zhao J, Lu W, Deng D (2017) Gibberellin in plant height control: old player, new story. Plant Cell Rep 36: 391-398. PMid:28160061
View Article PubMed/NCBIAchard P, Genschik P (2009) Releasing the brakes of plant growth: how GAs shutdown DELLA proteins. J Exp Bot 60: 1085-1092. PMid:19043067
View Article PubMed/NCBIAchard P, Gusti A, Cheminant S, Alioua M, Dhondt S, et al. (2009) Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr Biol 19: 1188-1193. PMid:19576768
View Article PubMed/NCBIOikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M (2004) A role of OsGA20ox1 , encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol 55: 687-700. PMid:15604710
View Article PubMed/NCBIJi SH, Gururani MA, Lee JW, Ahn BO, Chun SC (2014) Isolation and characterisation of a dwarf rice mutant exhibiting defective gibberellins biosynthesis. Plant Biol (Stuttg) 16: 428-439. PMid:23944972
View Article PubMed/NCBILuo Y, Dong X, Yu T, Shi X, Li Z, et al. (2015) A Single Nucleotide Deletion in Gibberellin20-oxidase1 Causes Alpine Dwarfism in Arabidopsis. 168: 930-937. PMid:25941313
View Article PubMed/NCBIHuang R, Li Y, Tang G, Hui S, Yang Z, et al. (2018) Dynamic phytohormone profiling of rice upon rice black-streaked dwarf virus invasion. J Plant Physiol 228: 92-100. PMid:29886196
View Article PubMed/NCBITang J, Han Z, Chai J (2016) Q&A: what are brassinosteroids and how do they act in plants? BMC Biol 14: 113. PMid:28007032
View Article PubMed/NCBITong H, Xiao Y, Liu D, Gao S, Liu L, et al. (2014) Brassinosteroid regulates cell elongation by modulating gibberellin metabolism in rice. Plant Cell 26: 4376-4393. PMid:25371548
View Article PubMed/NCBIAli S, Ganai BA, Kamili AN, Bhat AA, Mir ZA, et al. (2018) Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol Res 212-213: 29-37. PMid:29853166
View Article PubMed/NCBISels J, Mathys J, De Coninck BM, Cammue BP, De Bolle MF (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol Biochem 46: 941-950. PMid:18674922
View Article PubMed/NCBIAgrawal GK, Jwa NS, Rakwal R (2000) A novel rice (Oryza sativa L.) acidic PR1 gene highly responsive to cut, phytohormones, and protein phosphatase inhibitors. Biochem Biophys Res Commun 274: 157-165. PMid:10903912
View Article PubMed/NCBIAgrawal GK, Rakwal R, Jwa NS (2000) Rice (Oryza sativa L.) OsPR1b gene is phytohormonally regulated in close interaction with light signals. Biochem Biophys Res Commun 278: 290-298. PMid:11097833
View Article PubMed/NCBIFinkina EI, Melnikova DN, Bogdanov IV, Ovchinnikova TV (2017) Plant Pathogenesis-Related Proteins PR-10 and PR-14 as Components of Innate Immunity System and Ubiquitous Allergens. Curr Med Chem 24: 1772-1787. PMid:27784212
View Article PubMed/NCBIZhang D, Liang W, Yin C, Zong J, Gu F, et al. (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149-162. PMid:20610705
View Article PubMed/NCBI