Jin-Ying Gou
Email: jygou@fudan.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
Jin-Ying Gou
Email: jygou@fudan.edu.cn
Hao-Jie Sun1, Chu-Yang Wang1, Guo-Liang Zhang1, Jin-Ying Gou1
1State Key Laboratory of Genetic Engineering, MOE Key Laboratory for Biodiversity Science and Ecological Engineering, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai 200438, China.
Zhao-Shi Xu(xuzhaoshi@caas.cn)
Zhifen Pan(panzf@cib.ac.cn)
Ruihui Wang(wangrh@hebau.edu.cn)
Miguel Lara(mflara@ibt.unam.mx)
Hao-Jie Sun, Chu-Yang Wang, Guo-Liang Zhang, Jin-Ying Gou, Genomic analyses of wheat Aspartic proteinase gene family provide novel insights for wheat stress responses(2020)Journal of Plant Science 4(1) p:174-185
Aspartic proteinases (APs) are widely involved in a variety of physiological processes in plants, including development, stress resistance, and senescence. The biochemical properties of APs endow them many useful application prospects in both industrial production and scientific research, but there is little progress in wheat, an essential food crop. Two hundred thirty-seven members of the AP gene family were detected in wheat, significantly bigger than those in Arabidopsis or rice. The homology and gene structure of APs showed significant differences among AP subfamilies, as well as in the number of introns. Surprisingly, at the whole gene-family level, APs were significantly induced by phosphorus starvation, suppressed by heat stress, but not regulated by cold stress. After infection by Septoria tritici, APs were first suppressed but then gradually induced, and induced by Fusarium graminearum at four days post-inoculation (DPI), and young leaves infected by Septoria tritici at 13 DPI. For biotrophic pathogens, APs gradually increased after stripe rust infection, eventually up to 3-folds at whole gene family level at 11 DPI. This work provided useful information for APs in both abiotic and biotic stresses in wheat. The differentially expressed genes could serve as promising targets in future studies of wheat stress responses.
Keywords: Aspartic proteinase; wheat; transcriptome; stress; fungal response
Aspartic proteinases (Aps, EC3.4.23) widely exist in monocotyledonous and dicotyledonous plants [1]. Most APs are most active at acidic pH and inhibited by Pepstatin A [2]. The active site of typical APs contained two active Asp residues: Asp32 and Asp215 under X-ray diffraction crystallography [3].
HvAP (later renamed as phytepsin) was isolated from barley grains with an optimum pH of 3.5, with cDNA of 1863 bp, which encoded a 66 amino acids (a.a.) presegment and a mature protein with 442 a.a. including a 104-a.a. barley-specific region [4]. Structural analysis of barley AP proenzyme showed that the PSI fragment formed an independent domain, and besides prosegment, the presence of 13 residues at the N-terminal was also essential to keep the AP proenzyme inactive [5]. The mature form of phytepsin was a heterologous dimer 40kDa (29+11) and came from a precursor 48kDa (32+16) after processing. During its transportation into vacuoles, the primary translation product of phytepsin was first cleaved off N-terminal signaling peptide, probably 25 a.a. residues [6]. The cleavage of phytepsin occurs in the late-Golgi compartment or post-Golgi compartment [7]. The immature precursor of HvAP was likely bound to the receptor on the Golgi body membrane during sorting, which was then packaged and transported to the vesicle, and finally located in the vacuole [5]. Another study showed that barley AP Phytepsin detached from the ER via the COPII-mediated pathway and eventually entered the vacuole, which was not affected by the absence of the PSI domain [8].
In the Arabidopsis thaliana genome, 51 genes encode APs and fall into three categories: typical AP, nucellin-like AP, and atypical AP. Most APs have a typical signal of secretory system, with few bound with membrane system, eight in chloroplast, and two in mitochondrial, suggesting different physiological functions of APs in plants [9]. Analyses of the genome database of rice revealed that 96 predictive genes encoded AP and widely expressed in various tissues of rice [10]. Two atypical APs named nepenthesin I and II were isolated from the pitcher fluids of Nepenthes distillatoriatoria, with the maximum activity at pH2.6. These kinds of APs are very stable and have a high percentage of cysteine residues in their amino acid sequence. A bioinformatic study predicted that this AP could form six unique disulfide bonds, explaining its stability at 50 degrees [11]. Another in vitro study showed that recombinant nepenthesin-1 had the optimal pH of 2.5 and was very stable at high pH, but less stable than nepenthesin-1 extracted in vivo, possibly due to lacking glycosides modification [12]. The nepenthesin subfamilies share only 20% homology with typical plant APs and have a nepenthesin-type AP-specific insertion different from typical PSI [12].
APs involve in plants’ resistance to biotic stresses. Aspartic proteases in Arabidopsis (AT2G17760) and Vitis vinifera (TC49155) increased their expression levels in Arabidopsis inoculated with TMV-Cg and Vitis vinifera inoculated with grapevine leafroll-associated virus strain 3 (GLRaV-3) [13]. The co-expression with senescence-associated genes suggested that APs could take part in the nutrients mobilization, protein degradation, and other physiological processes related to the senescence [13]. Overexpression of CDR1, a gene encoding apoplastic AP in Arabidopsis, resulted in dwarfism and increased resistance to Pseudomonas syringae. While loss-of-function mutation of CDR1 led to susceptibility to P. syringae compared with the wild type, during infection, CDR1 accumulated in the intercellular spaces and produced a mobile signal peptide, which can be inhibited by Pepstatin A and enzymatic site mutation of CDR1 [14]. BAG6 was essential in the basic immunity of Arabidopsis thaliana against the fungus B.cinerea, and APCB1 encodes an AP protein that cleaves BAG6, thus to activate downstream autophagy responses to resist pathogens [15]. In brief, these above studies suggested that AP might play a role in resistance pathway in an indirect way.
APs are also associated with plant resistance to abiotic stresses. After drought treatment, the transcription and the activity of APs significantly increased in drought-susceptible beans [16]. An AP member, ASPG1 (ASP in Guard cell 1), was up-regulated in drought stress in potato [17]. FeAP9, a typical AP in buckwheat, was increased at expression level in buckwheat leaves subjected to abiotic stresses (darkness, drought, and UV-B), injuries, and SA treatment [18]. The expression of plant aspartic proteinase A3 (PASPA3) was increased 19 fold in maize under waterlogged conditions [19]. The above results suggested the involvement of APs in the process of plant resistance to abiotic stress.
Compared with the extensive researches of APs in model plants such as Arabidopsis and rice, there is little progress in wheat, one of the most critical food crops feeding 70% of people in the world. The production of wheat is strongly affected by biotic and abiotic stresses, in which APs could play essential roles. In this work, we analyzed the gene family of APs in wheat using the latest genome information. Then the expression patterns of APs in 72 tissues along with development were analyzed. Moreover, expression patterns of APs showed significant variations in abiotic and biotic stresses, suggesting their essential roles in those treatments. These results could provide beneficial information for future study of APs in wheat.
2.1 Genome-wide identification of AP family genes in wheat
The latest version (CS1.1) of IWGSC wheat genome information was from Ensembl Plants (http://plants.ensembl.org/Triticum_aestivum/) [20]. We identified two hundred fifty-five putative genes coding for aspartic proteinase using the keyword "aspartic proteinase." After removing different transcripts of the same genes, 237 APs had unique gene identifiers. The genome sequence, coding sequences (CDS), and detailed chromosome locations of each gene were downloaded from Ensemble Plants using the gene identifiers acquired above. We first calculated isoelectric point (pI) and molecular weight (Mw) of each protein in ExPASy (http://web.expasy.org/compute_pi/). We next predicted the subcellular localizations through Target1.1 Server, ChloroP1.1 Server, and SignalP5.0 [21]. The genome sequences and CDS of each gene were then uploaded to the Gene Structure Display website to draw the gene structures and calculate numbers of introns. The Gene identifier, for example, TraesCS3B02G269300, in which "CS” is the abbreviation of Chinese Spring, “3B” means the third chromosome of B genome, 02G means version1.1 and 269300 standards for the location in the chromosome.
2.2 Phylogenetic analyses and expression of APs in wheat
The amino acid sequences of 237 aspartic proteinases were downloaded from Ensemble Plants (http://plants.ensembl.org/Triticum_aestivum/) and used in the phylogenetic analysis. We constructed the phylogenetic tree using the MEGA X software using the neighbor-joining method with the option of 500 replicates of bootstrap.
The expression data of each aspartic proteinases were from the Wheat eFP Browser (http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi). There were 72 samples in total, but only 13 tissues worked as representatives in the figure. We produced the heatmap using the logarithms of the original PTM values through Graphpad Prism 8 software.
2.3 Expression of APs under stresses
RNA-Seq data were from the Wheat expression Browser (http://www.wheat-expression.com/), including all the abiotic and biotic stress [22]. Expression data of the 237 aspartic proteinases were extracted in the form of primary TPM values and then transformed into log values, and draw the heatmap in Graphpad Prism 8. The results were processed and refined using Adobe Illustrator. We further calculated the significance between control and the treated samples in each experiment using paired t-test in R.
2.4 Conserved Motifs analysis of APs.
The amino acid sequences of 237 aspartic proteinases were analyzed on the Multiple Expectation Maximization for Motif Elicitation (MEME) web server to identify the conserved motifs. Five motifs with high scores were selected to draw a binary heatmap in Graphpad prism 8. The results were refined and processed using Adobe illustrator.
2.5 Cis-acting elements analysis in the promotors of APs.
We used the 2.5kb UTR sequences of each aspartic proteinases to identify Cis-acting element through Plant CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The numbers of nine stress-related cis-acting elements in the promotors of 237 APs constituted a table and generated a binary heatmap through Graphpad prism 8. The results were refined and processed using Adobe illustrator. We further investigated the correlation between the numbers of cis-element and expression level (TPM values) under stresses using paired t-test in R.
3.1 Identification of AP gene family in wheat
Based on the recently released IWGSC wheat genome annotation, we searched the wheat genome for AP gene family members using the keywords “Aspartic proteinase.” In total, 237 putative APs were identified (Supplemental Table S1). The significant increases of AP family genes in wheat compared with Arabidopsis or rice suggest that they are likely to play essential roles in the developmental and physiological processes of wheat. We next calculated isoelectric point (pI) and molecular weight (Mw) of each AP and predicted subcellular localization of each AP (Supplemental Table S1). One hundred forty-one APs contained secretion signal peptides, and 37 APs were likely chloroplast proteins, most of which were from Aspartic proteinase nepenthesin-1 (APn1) and Aspartic proteinase nepenthesin-2 (APn2) classes.
Based on sequence annotations, the above 237 APs formed the following major groups: APn1, APn2, Aspartic proteinase (Asp1), Nucellin-like aspartic protease (NAP), Aspartic proteinase (Asp2) and ASPARTIC PROTEASE IN GUARD CELL 1 (APG1). Among them, the APn1 class has 131 members and represents the most abundant class. The APn2 class has 48 members and represents the second one.
We performed a phylogenetic analysis of the 237 members of APs in wheat with their amino acid sequences using the NJ method. The phylogenetic distribution agreed with the genome annotation. The phylogenetic tree showed that the AP gene family composed six main classes, including nine subclasses (Fig. S1). In the phylogenetic tree, Asp1, Asp2, and NAP were close to each other and formed an “Asp” subfamily.
The APn1 class has 33 members in chromosome 2, accounting for 86.8% of the total aspartic proteinase family in chromosome 2, which is a very significant enrichment (hypergeometric distribution, p < 0.0001). Moreover, in chromosome 1, there are six Asp class genes, accounting for 2/3 of the total, which represents a significant enrichment (hypergeometric distribution, p < 0.001). As the A, B, and D genomes each have 77, 81, and 72 AP members, respectively, there is no apparent difference among them in our analysis.
3.2 Expression of APs during wheat development
We next studied expression patterns of the whole AP family genes using the RNAseq data downloaded from the wheat eFP Browser, including 72 tissues at different developmental stages (Supplementary Table S2). Among them, we selected 13 representative tissues and stages to produce a heatmap to illustrate the expression patterns of all the 237 APs (Supplemental Figure 1). The expression levels of most APn1 and APn2 class genes were under detectable level in most tissues, with only several APn1 class genes expressed in grain at the Milk grain stage, suggesting their potential functions in grain development (Fig. 1). The Asp class genes usually have higher expression levels in multiple tissues during the whole life cycle of wheat (Figure 1). Notably, among whole wheat developmental stages in various tissues, TraesCS1B02G434000, TraesCS1D02G412200, and TraesCS1A02G404500 have the highest expression, and they are homeologs of the same gene in different genomes. The expression pattern suggested that they could play a dominant role in AP family and are therefore named TaAP-1A, TaAP-1B, and TaAP-1D.
Gene structures of each gene have apparent differences between different subfamilies. The intron numbers of all the Asp class genes are over 6. Nevertheless, intron numbers of all the APn1 and APn2 class genes are less than two. The majority of APn1 class genes even have no intron at all (Fig. 1). The above pattern of the gene structures is consistent with their distribution in the phylogenetic tree, suggesting that each subfamily could evolve independently.
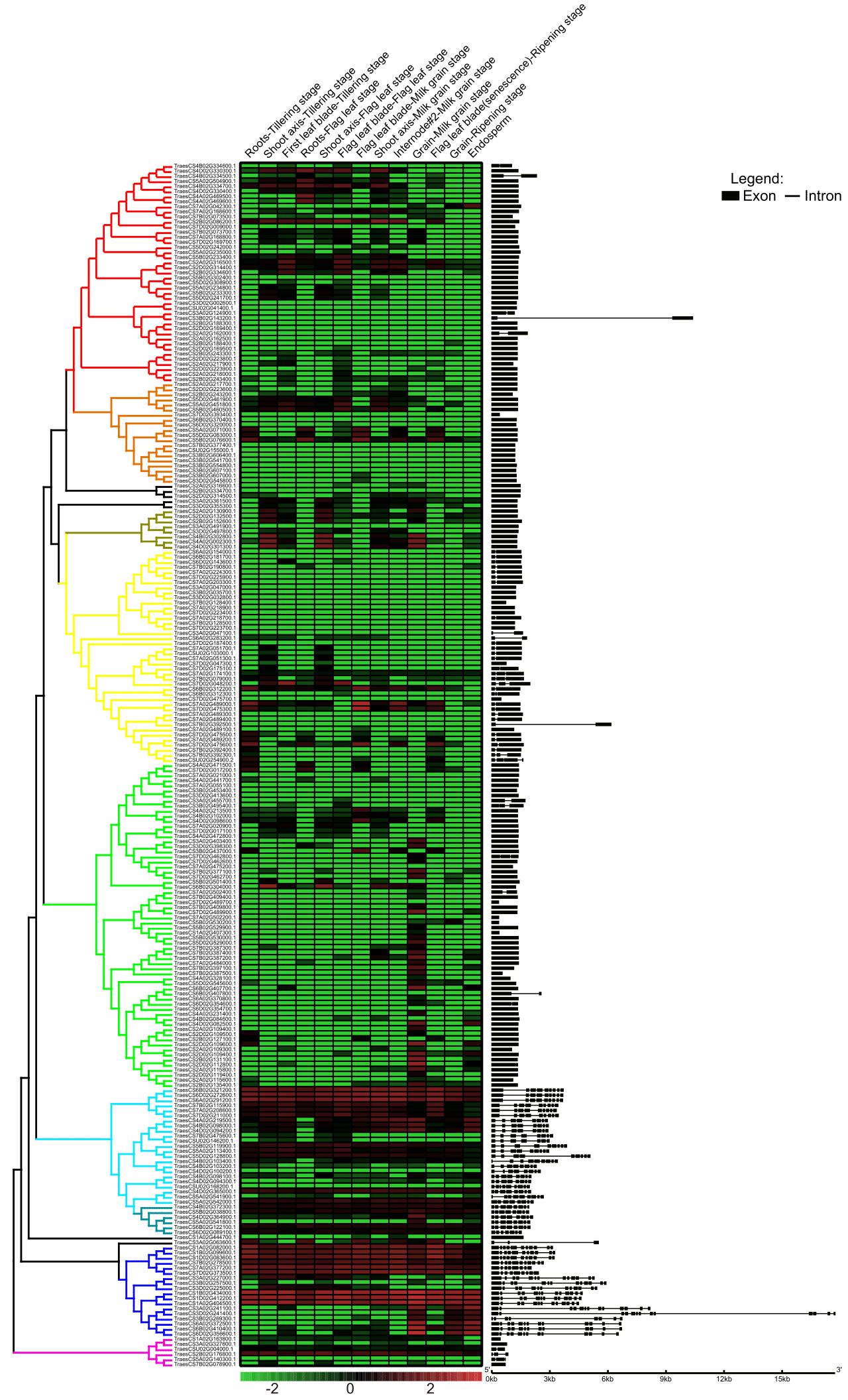
Fig. 1 Phylogenetic, expression pattern, and gene structure analyses of wheat AP family genes. Aspartic proteinase nepenthesin-1: red, orange, and green. Aspartic proteinase nepenthesin-2: yellow. Aspartic proteinase Asp1: cyan. Nucellin-like: dark cyan. Aspartic proteinase: blue. Unclassified: pink.
3.3 Expression of AP family genes under abiotic and biotic stresses
After collecting information in developmental stages and tissues, we then compared the expression patterns of APs in abiotic stresses (Supplementary Table S3, Fig. 2A). After cold treatment at three-leaf stages, the expression of 7 APs increased, while another seven decreased in shoots, with no significant change in the overall APs (Students’ paired t-test p = 0.32). In contrast, upon heat treatment for one hour, only 2 AP genes increased while another 26 decreased their expression levels, which was a very significant decrease at overall APs (Student paired t-test p = 0.0027) (Supplementary Table S4, Fig. 2B). These data suggested that APs could confer different biological functions between heat and cold stresses in wheat. The overall expression of APs did not exhibit any significant changes after PEG6000 treatment at 2 and 12 hours, although a few low expression genes further decreased. After phosphorus starvation, APs were induced at the whole family level both in roots and shoots at 24 hours (Fig. 2C), suggesting that APs could very likely involve in either the acquisition of environmental phosphorus or remobilization of endogenous phosphorus flux to essential biological events.
We next checked the expression of APs in biotic stresses, which caused 15~35% decrease in wheat production and led to billion-dollar losses [23]. Septoria tritici infection is the cause of glume blotch of wheat, and it gradually induced the expression of wheat APs from 4 to 13 days post-inoculation (DPI), which was a significant increase at 13 DPI. The expression of 13 genes increased, while another 25 decreased in rachis upon F. graminearum inoculation, with around 60% reduction of total APs. The above change represented a significant reduction (Student paired t-test p < 0.05), suggesting that F. graminearum could suppress the function of AP in the infection process (Supplementary Table S4). One AP (TraesCS4B02G102000.1), with a very significant increase in the RNAseq analysis, matched a probe in the Affymetrix wheat 61k microarray (TaAffx.21249.1.S1), which exhibited a 10-fold increase at four days post F. graminearum inoculation [24].
Different biotrophic pathogens also regulated the expressions of APs. After the inoculation of stripe rust 87/66, the overall signal of APs showed significant increases from 3DPI and further increased along with the pathogenesis, resulted over 2-fold increase at 11 DPI (Students’ paired t-test p < 0.01; Fig. 2D). Similarly, upon inoculation of powdery mildew E09, an increase showed up in the AP gene family (Students’ paired t-test p < 0.01) at 2DPI. The above differential expression suggested that some APs could very likely involve in wheat responses to fungal pathogens.
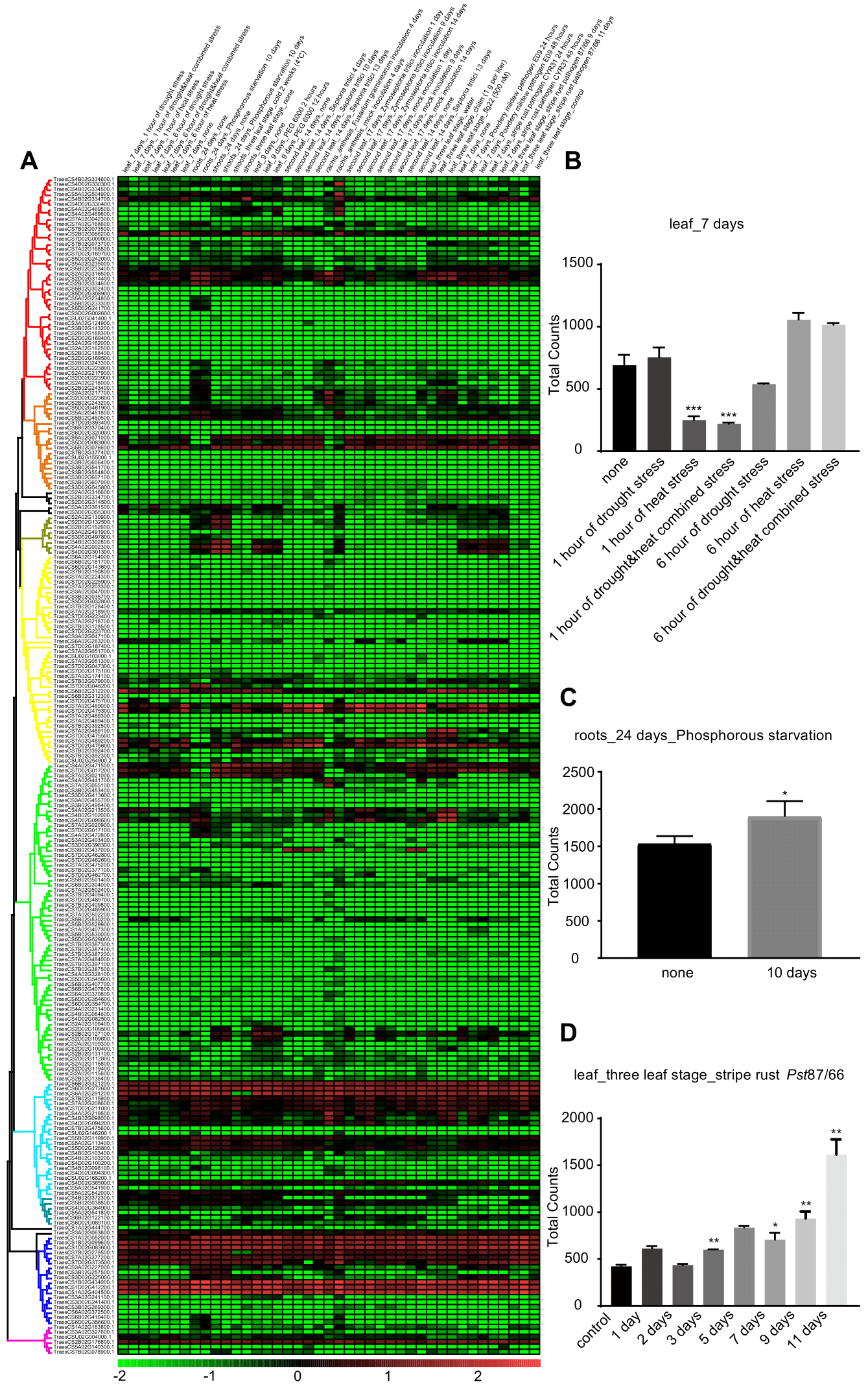
Fig.2 The stress responses of APs. (A) The expression patterns of APs in different stress treatments. The normalized logarithm of TPM values were used to produce the heatmap. The column identifier presented tissue, age, and stress. (B) The overall expression of APs in leaves treated by heat and/or drought. (C) The overall signal of APs in roots after phosphorylation starvation for ten days. (D) The overall signal of APs in leaves inoculated with stripe rust pathogen. Data represented three biological repeats. Students’ paired t-test, * p < 0.05, ** p < 0.01.
3.4 Conserved Motifs and Domains on APs
Conserved motifs in the APs were identified based on their amino acid sequences on the MEME web server. Five conserved motifs were identified (Supplementary Table S5). Motif 1 and motif 2 exist in most of APs, while motif 3 only exists in APn1 and APn2 subfamilies. This specificity of APn1 and APn2 anastomosed with the low intron numbers in their genome structure (Fig. 3). It is worth mentioning that, motif 4 and motif 5 are unique to the Asp class, especially those with the highest expression level among all the whole AP gene family. The amino acid residues DTG on motif5 and DSG on motif1 together constituted the catalytic sites DTG/DSG of the typical aspartic proteinases [25].
Six genes on the bottom of the phylogenetic tree, however, have no common conserved motifs at all. Future AP biochemical activity analyses of these APs would contribute to improve the accuracy of wheat genome annotation or increase our understanding of AP.

Fig. 3 Conserved motifs in APs. Purple means the existence of the conserved motif, while orange means missing of the corresponding motif in the protein.
3.5 Distribution of Cis-Acting elements in AP promoters
Cis-acting elements in promoters are the anchor sites for regulatory factors, transcription factors, for example. We downloaded 2.5kb sequences upstream each 237 APs and analyzed them through the plant CARE webserver to identify potential cis-acting elements. To explore the correlation between gene expression and the distribution of cis-acting elements, we analyzed nine stress-response related cis-acting elements, including TC-rich repeats (defense and stress) [26], LTR (low- temperature response) [27], TCA-element(salicylic acid response) [28], ABRE (abscisic acid response) [29], CGTCA-motif (MeJA-response) and TGACG-motif (MeJA-response) [30], P-box (gibberellin-response) [31], GARE-motif(gibberellin-response) [32] , TATC-box(gibberellin-response) [33], and MBS (drought-inducible MYB binding site) [34] (Fig. 4; Supplementary Table S6).
Using Person’s test, we found that the number of TATC-Box had a significant correlation with the expression levels of APs under the treatment of drought and heat. The numbers of TCA-element are significantly correlated with expression levels of APs under the cold stress. The numbers of GARE-motif significantly correlated with the expression levels of APs under treatments of drought and heat, chitin, cold stress, Fusarium graminearum inoculation, and multiple experiments using three-leaf-stage leaves regardless of being treated or not (Supplementary Table S7). These results showed that APs play a part in the developmental processes in wheat and are regulated by phytohormone such as gibberellin.
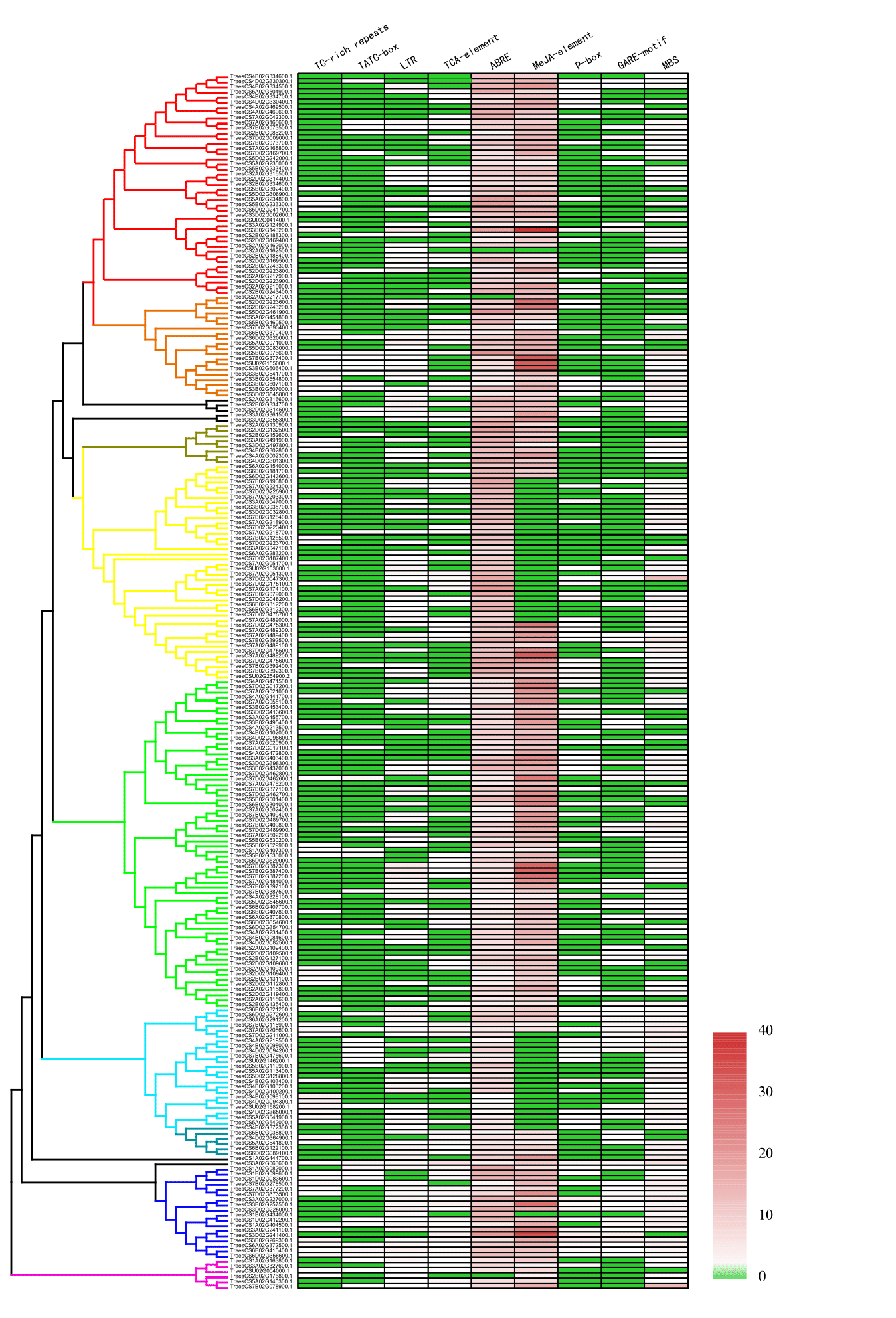
Fig. 4 Cis-Acting elements in promoters of AP gene family. Nine stress-related cis-acting elements were presented. The numbers of elements varies from 0 to 40, which were colored from green to red. TC-rich repeats: cis-acting element involved in defense and stress responsiveness. TATC-box: cis-acting element involved in gibberellin-responsiveness. LTR: cis-acting element involved in low-temperature responsiveness. TCA-element: cis-acting element involved in salicylic acid responsiveness. ABRE: cis-acting element involved in the abscisic acid responsiveness. P-box: gibberellin-responsive element. GARE-motif: gibberellin-responsive element. MBS: MYB binding site involved in drought.
Wheat AP genes form different subfamilies conserved in both gene structures and functional motifs. The expression variations among different APs during development, environmental stresses, and biological stresses suggested that certain APs could play essential roles in corresponding biological processes.
To Download All Files : Click Here
Supplementary Table S1. AP gene family in the genome of Chinese Spring.
Supplementary Table S2. Expression of APs during wheat development.
Supplementary Table S3. Expression of APs under stresses.
Supplementary Table S4. Paired student's t-test between control and stressed samples.
Supplementary Table S5. Conserved motifs in APs.
Supplementary Table S6. Cis-acting elements in AP promoters.
The authors gratefully acknowledge financial supports provided by the National Natural Science Foundation of China (31772146 and 31972350).
Conflicts of interest
The authors do not have any conflicts of interest to disclose.
G. Voigt, B. Biehl, H. Heinrichs, J. Voigt, Aspartic proteinase levels in seeds of different angiosperms, Phytochemistry 44 (1997) 389-392. 00485-2
View ArticleI. Simoes, C. Faro, Structure and function of plant aspartic proteinases, Eur J Biochem 271 (2004) 2067-2075. PMid:15153096
View Article PubMed/NCBIL. Pearl, T. Blundell, The active site of aspartic proteinases, FEBS Letters 174 (1984) 96-101. 81085-6
View ArticleP. Runeberg-Roos, K. Tormakangas, A. Ostman, Primary structure of a barley-grain aspartic proteinase. A plant aspartic proteinase resembling mammalian cathepsin D, Eur J Biochem 202 (1991) 1021-1027. PMid:1722454
View Article PubMed/NCBIJ. Kervinen, G.J. Tobin, J. Costa, D.S. Waugh, A. Wlodawer, A. Zdanov, Crystal structure of plant aspartic proteinase prophytepsin: inactivation and vacuolar targeting, EMBO J 18 (1999) 3947-3955. PMid:10406799
View Article PubMed/NCBIP. Runeberg-Roos, J. Kervinen, V. Kovaleva, N.V. Raikhel, S. Gal, The aspartic proteinase of barley is a vacuolar enzyme that processes probarley lectin in vitro, Plant Physiol 105 (1994) 321-329. PMid:8029356
View Article PubMed/NCBIS. Glathe, J. Kervinen, M. Nimtz, G.H. Li, G.J. Tobin, T.D. Copeland, D.A. Ashford, A. Wlodawer, J. Costa, Transport and activation of the vacuolar aspartic proteinase phytepsin in barley (Hordeum vulgare L.), J Biol Chem 273 (1998) 31230-31236. PMid:9813030
View Article PubMed/NCBIK. Tormakangas, J.L. Hadlington, P. Pimpl, S. Hillmer, F. Brandizzi, T.H. Teeri, J. Denecke, A vacuolar sorting domain may also influence the way in which proteins leave the endoplasmic reticulum, Plant Cell 13 (2001) 2021-2032. PMid:11549761
View Article PubMed/NCBIC. Faro, S. Gal, Aspartic proteinase content of the Arabidopsis genome, Curr Protein Pept Sci 6 (2005) 493-500. PMid:16381599
View Article PubMed/NCBIJ. Chen, Y. Ouyang, L. Wang, W. Xie, Q. Zhang, Aspartic proteases gene family in rice: Gene structure and expression, predicted protein features and phylogenetic relation, Gene 442 (2009) 108-118. PMid:19409457
View Article PubMed/NCBIK. Takahashi, S.B. Athauda, K. Matsumoto, S. Rajapakshe, M. Kuribayashi, M. Kojima, N. Kubomura-Yoshida, A. Iwamatsu, C. Shibata, H. Inoue, Nepenthesin, a unique member of a novel subfamily of aspartic proteinases: enzymatic and structural characteristics, Curr Protein Pept Sci 6 (2005) 513-525. PMid:16381601
View Article PubMed/NCBIA. Kadek, V. Tretyachenko, H. Mrazek, L. Ivanova, P. Halada, M. Rey, D.C. Schriemer, P. Man, Expression and characterization of plant aspartic protease nepenthesin-1 from Nepenthes gracilis, Protein Expr Purif 95 (2014) 121-128. PMid:24365662
View Article PubMed/NCBIC. Espinoza, C. Medina, S. Somerville, P. Arce-Johnson, Senescence-associated genes induced during compatible viral interactions with grapevine and Arabidopsis, J Exp Bot 58 (2007) 3197-3212. PMid:17761729
View Article PubMed/NCBIY. Xia, H. Suzuki, J. Borevitz, J. Blount, Z. Guo, K. Patel, R.A. Dixon, C. Lamb, An extracellular aspartic protease functions in Arabidopsis disease resistance signaling, EMBO J 23 (2004) 980-988. PMid:14765119
View Article PubMed/NCBIY. Li, M. Kabbage, W. Liu, M.B. Dickman, Aspartyl Protease-Mediated Cleavage of BAG6 Is Necessary for Autophagy and Fungal Resistance in Plants, Plant Cell 28 (2016) 233-247. PMid:26739014
View Article PubMed/NCBIM.H. Cruz de Carvalho, A. d'Arcy-Lameta, H. Roy-Macauley, M. Gareil, H. El Maarouf, A.T. Pham-Thi, Y. Zuily-Fodil, Aspartic protease in leaves of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata L. Walp): enzymatic activity, gene expression and relation to drought susceptibility, FEBS Lett 492 (2001) 242-246. 02259-1
View ArticleL. Gong, H. Zhang, X. Gan, L. Zhang, Y. Chen, F. Nie, L. Shi, M. Li, Z. Guo, G. Zhang, Y. Song, Transcriptome Profiling of the Potato (Solanum tuberosum L.) Plant under Drought Stress and Water-Stimulus Conditions, PLoS One 10 (2015) e0128041. PMid:26010543
View Article PubMed/NCBIG.S. Timotijevic, M. Milisavljevic, S.R. Radovic, M.M. Konstantinovic, V.R. Maksimovic, Ubiquitous aspartic proteinase as an actor in the stress response in buckwheat, J Plant Physiol 167 (2010) 61-68. PMid:19643510
View Article PubMed/NCBIK. Arora, K.K. Panda, S. Mittal, M.G. Mallikarjuna, A.R. Rao, P.K. Dash, N. Thirunavukkarasu, RNAseq revealed the important gene pathways controlling adaptive mechanisms under waterlogged stress in maize, Sci Rep 7 (2017) 10950. PMid:28887464
View Article PubMed/NCBIE. Pennisi, Detailed genome maps paths to better wheat, Science 361 (2018) 635. PMid:30115790
View Article PubMed/NCBIO. Emanuelsson, S. Brunak, G. von Heijne, H. Nielsen, Locating proteins in the cell using TargetP, SignalP and related tools, Nat Protoc 2 (2007) 953-971. PMid:17446895
View Article PubMed/NCBIR.H. Ramirez-Gonzalez, P. Borrill, D. Lang, S.A. Harrington, J. Brinton, L. Venturini, M. Davey, J. Jacobs, F. van Ex, A. Pasha, Y. Khedikar, S.J. Robinson, A.T. Cory, T. Florio, L. Concia, C. Juery, H. Schoonbeek, B. Steuernagel, D. Xiang, C.J. Ridout, B. Chalhoub, K.F.X. Mayer, M. Benhamed, D. Latrasse, A. Bendahmane, B.B.H. Wulff, R. Appels, V. Tiwari, R. Datla, F. Choulet, C.J. Pozniak, N.J. Provart, A.G. Sharpe, E. Paux, M. Spannagl, A. Brautigam, C. Uauy, The transcriptional landscape of polyploid wheat, Science 361 (2018). PMid:30115782
View Article PubMed/NCBIW. Chen, C. Wellings, X. Chen, Z. Kang, T. Liu, Wheat stripe (yellow) rust caused by Puccinia striiformis f. sp. tritici, Mol Plant Pathol 15 (2014) 433-446. PMid:24373199
View Article PubMed/NCBIP.-F. Qi, A. Johnston, M. Balcerzak, H. Rocheleau, L.J. Harris, X.-Y. Long, Y.-M. Wei, Y.-L. Zheng, T. Ouellet, Effect of salicylic acid on Fusarium graminearum, the major causal agent of fusarium head blight in wheat, Fungal Biology 116 (2012) 413-426. PMid:22385623
View Article PubMed/NCBIY. Wan, R.L. Poole, A.K. Huttly, C. Toscano-Underwood, K. Feeney, S. Welham, M.J. Gooding, C. Mills, K.J. Edwards, P.R. Shewry, R.A. Mitchell, Transcriptome analysis of grain development in hexaploid wheat, BMC Genomics 9 (2008) 121. PMid:18325108
View Article PubMed/NCBIF. Diaz-De-Leon, K.L. Klotz, L.M. Lagrimini, Nucleotide sequence of the tobacco (Nicotiana tabacum) anionic peroxidase gene, Plant Physiol 101 (1993) 1117-1118. PMid:8310051
View Article PubMed/NCBIM.A. Dunn, A.J. White, S. Vural, M.A. Hughes, Identification of promoter elements in a low-temperature-responsive gene (blt4.9) from barley (Hordeum vulgare L.), Plant Mol Biol 38 (1998) 551-564. PMid:9747801
View Article PubMed/NCBIA.P. Goldsbrough, H. Albrecht, R. Stratford, Salicylic acid-inducible binding of a tobacco nuclear protein to a 10 bp sequence which is highly conserved amongst stress-inducible genes, Plant J 3 (1993) 563-571. PMid:8220463
View Article PubMed/NCBIT. Hobo, M. Asada, Y. Kowyama, T. Hattori, ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent, Plant J 19 (1999) 679-689. PMid:10571853
View Article PubMed/NCBIJ. Rouster, R. Leah, J. Mundy, V. Cameron-Mills, Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain, Plant J 11 (1997) 513-523. PMid:9107039
View Article PubMed/NCBIK. Itoh, J. Yamaguchi, N. Huang, R.L. Rodriguez, T. Akazawa, K. Shimamoto, Developmental and Hormonal Regulation of Rice [alpha]-Amylase(RAmy1A)-gusA Fusion Genes in Transgenic Rice Seeds, Plant Physiol 107 (1995) 25-31. PMid:12228339
View Article PubMed/NCBIJ.K. Kim, J. Cao, R. Wu, Regulation and interaction of multiple protein factors with the proximal promoter regions of a rice high pI alpha-amylase gene, Mol Gen Genet 232 (1992) 383-393. PMid:1375314
View Article PubMed/NCBIF. Gubler, R. Kalla, J.K. Roberts, J.V. Jacobsen, Gibberellin-regulated expression of a myb gene in barley aleurone cells: evidence for Myb transactivation of a high-pI alpha-amylase gene promoter, Plant Cell 7 (1995) 1879-1891. PMid:8535141
View Article PubMed/NCBIT. Urao, K. Yamaguchi-Shinozaki, S. Urao, K. Shinozaki, An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence, Plant Cell 5 (1993) 1529-1539. PMid:8312738
View Article PubMed/NCBI