Lan Jing
Email: jinglan71@126.com
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 5 ISSUE: 3
Page No: 631-640
Lan Jing
Email: jinglan71@126.com
Yan Lu, Yang Song, Xinchun Li, Meng Zhang, Lan Jing*
College of Horticulture and Plant Protection, Inner Mongolia Agricultural University, Hohhot 010011, China
Zhang Z(biozhang0713@163.com)
Yan Lu, Yang Song, Xinchun Li, Meng Zhang, Lan Jing, The characteristic of a novel metalloproteinase from Puccinia helianthin (2021) Journal of Computational Chemistry & Molecular Modeling 5(3) p:631-640
In phytopathogenic fungi, several metalloproteinases have been implicated in virulence, but pathogenesis roles in rust are unknown. Here, we identified a M36 family metalloproteases from Puccina helianthi genome, designated PhMEP1, aimed to reveal the biological characteristics related to pathogenicity. Bioinformatics analysis showed that PhMEP1contains some of the conserved zinc-binding and active site motifs characteristics of metalloproteinase which might well be a new fungalysin-like peptidase in the M36 family. The PhMEP1 protein was overexpressed as inclusion bodies in Escherichia coli. The recombinant expression products of PhMEP1 could hydrolyze azocasein and the hydrolytic activity was suppressed by metal ion chelating agents such as 1,10-phenanthroline and ethylene diamine tetraacetic acid (EDTA). The subcellular localization of PhMEP1 was detected by confocal laser scanning microscopy of the leaves from a 2-day-old Nicotiana benthamiana seedlings expressed PhMEP1-GFP fusion that showed green fluorescent protein (GFP) signal accumulated in the plasma membrane of epithelial cells. PhMEP1 may serve as a pathogenicity factor contributing to P. helianthi penetration and infection.
Keywords: Metalloproteinase, Puccinia helianthi, M36 family, Hydrolytic activity, Subcellular localization
Sunflower rust, which is one of the major diseases of sunflower globally, spreads in the sunflower planting area triggering a heavy loss. The average prevalence rate in sunflower fields was more than 50%, in a rust epidemic year (Gulya et al., 2019). Sunflower rust is especially influenced by environmental conditions. when environmental conditions favor rust disease development the yield losses can be up to 80% (Markell et al., 2009). Breeding rust-resistant hybrids is considered the most effective, economical, easy to use, and environmentally friendly management for control of sunflower rust. Puccina helianthi, the causal agent of sunflower rust, is a macrocyclic and autoecious pathogen. To develop resistant plants, a better understanding of rust virulence factors, is needed. Exogenous absorption of macromolecular nutrients is a common strategy of fungal growth, which relies on the secretion of hydrolases including chitinases, cellulases, pectinases, and proteases. Of these proteins, metalloproteinases are a type of protease that contain conserved zinc-binding sites and HEXXH common sequences (Tallant et al., 2006; Zhou et al., 2018).
Metalloproteases of pathogens were reported have roles in virulence (Jia et al., 2000; Sanz-Martín et al., 2016). For instance, Avr-Pita, a zinc metalloprotease from the rice blast fungus Magnaporthe oryzae, recognized by a cytoplasmic resistance gene-encoded protein Pi-ta that triggers a signaling cascade leading to host rice resistance (Jia et al., 2000). Recently we have identified a M36 domain-containing metalloproteinase in P. helianthi, designated PhMEP1 (GenBank ID: KU994904), investigated its expression pattern strongly up-regulated during the early stages of infection (Jing et al., 2017; Li et al., 2019).
The first M36 metalloproteases was described from the opportunistic human pathogen Aspergillus fumigatus is involved in the degradation of proteinaceous structural barriers during invasion of the host lung (Monod et al., 1993). M36 metalloproteases from Fusarium fungi secrete fungalysins, that truncate maize class IV chitinases, to stop the chitinmediated activation of the plant immune system (Naumann et al., 2011). M36 metalloproteases from Trichophyton mentagrophytes is involved in the degradation of keratinase during invasion of the host skin (Zhang et al., 2014). M36 metalloproteases comes from A. fumigatus (Silva et al., 2013), Coprinopsis cinerea (Lilly et al., 2008), Perenniporia fraxinea (Kim et al., 2008), Trichophyton sp. (Jousson et al., 2004), Microsporum sp. (Jousson et al., 2004), Nomuraea atypicola (Ueda et al., 2013), Coccidioides posadasii (Sharpton et al., 2009), Pyrenophora tritici-repentis (NCBI protein XP 001940970.1), and Arthroderma gypseum (NCBI protein XP 003173813.1). In the present study, we analyzed the phylogenetic relationship of this metalloproteinase, and described the expression of the mature proteinase of PhMEP1in Escherichia coli. Moreover, the hydrolytic activity of this enzyme against azocasein was evaluated, and the subcellular localization of PhMEP1 was detected.
2.1. Rust pathogen isolates and sunflower
The urediospores of P. helianthi isolate SY used in the present study was produced by increasing on a universal susceptible varieties Heidapian.
2.2. Sequences analyses of full-length sequence of PhMEP1
The cDNA and genomic DNA was analyzed using BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi). SOPMA (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) was used to analysis the second structure and Phyre2 to predict the three-dimensions structure. The genome sequences and their corresponding annotated proteins database of the 20 MEPs are available from UniProt (www.uniprot.org), GenBank (http://www.ncbi.nlm.nih.gov/genbank/) and the Fungal Genome Initiative at the BROAD Institute (http://www.broad.mit.edu/annotation/fungi/fgi) databases. The phylogenetic tree was constructed by Mega 7.0 using neighbor-joining (parameters: 1000 bootstraps).
2.3. Cloning and heterologous expression of PhMEP1
To express the mature PhMEP1 in E. coli, MaxCodonTM Optimization Program (V13) was used to optimize the amino acid sequence of mature PhMEP1. Gene PhMEP1 was introduced to NdeI site and a HindIII site at the 5’ and 3’ ends, respectively, and a Met start codon was created by the NdeI site at the N terminus. Total genes were synthesized and inserted into expression vector Pet30a (His-tag at the N terminus). The clones with correct insertion were identified by enzyme digestion and sequencing. Pet30a-PhMEP1 was transformed into expression host strain E. coli BL21 (DE3). Positive colonies were identified by PCR and double digestion with NdeI and HindIII. The primers were designed with Primer Premier 5.0 program and synthesized by Invitrogen.
PhMEP1-F: 5’-CGCCATATGATGTCCTATCGCGTCTTTCCCTGGTCG-3’
PhMEP1-R: 5’-CCCAAGCTTTCAGTTCTCGCAAACACCTCTTGG-3’
PCR was conducted in 25 μL reaction system. The amplification protocol consisted of the denaturation at 95 ℃ for 5 min followed by 35 cycles with 30 s at 94 ℃, 30 s at 65.3 ℃, and 1 min at 72 ℃. A final elongation was done at 72 ℃ for 10 min. The cDNA obtained after the total RNA amplification was separated on a 1.5% agarose gel. A single colony of transformant clones containing Pet30a-PhMEP1 was then inoculated in 4 mL Luria-Bertani (LB) broth containing 50 µg mL-1 Kanamycin in a shaking incubator. When cells reached the log growth phase (OD600 0.5–0.8), isopropyl-β-D-thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM to induce production of T7 RNA polymerase and thus the synthesis of the PhMEP1, and cells were harvested after 16 h of growth at 15 ℃ or 37 ℃. The culture was then used for PhMEP1 expression analysis by Sodium Dodecyl Sulphate Poly Acrylamide Gel Electrophoresis (SDS-PAGE).
The transformant was cultured in 3 L LB medium. When cells reached the log growth phase (OD600 0.8), IPTG was added to induce PhMEP1 expression and the final concentration of IPTG was 1mM. The cells were harvested after 16 h of growth at 15 ℃.
2.4. Purification of PhMEP1
The cells were harvested by centrifugation (6000 rpm for 5 min at 4 ℃). The bacterial pellets were resuspended in lysis buffer [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM dithiothreitol (DTT), 20 mM imidazole containing 1% Triton X-100]. The bacterial cell pellets were lysed on ice with the aid of ultrasonic cell disruptor. At the same time, Ni-IDA affinity column was equilibrated with the buffer [50 mM Tris-HCl (pH 8.0), 300 mM NaCl, 20 mM imidazole]. The column was then eluted by the equilibrium buffer with different concentrations of imidazole (50 and 500 mM) to obtain the target protein. Elution components were collected for SDS-PAGE electrophoresis analysis.
The inclusion body was washed in buffer containing 50 mM Tris-HCl (pH 8), 300 mM NaCl containing 1% Triton X-100, 2 mM ethylene diamine tetraacetic acid (EDTA) and 5 mM DTT, and then resuspended in buffer containing 50 mM Tris-HCl (pH 8), 300 mM NaCl, 8 mM Urea, and 20 mM imidazole. At the same time, Ni-IDA affinity column was equilibrated with this buffer. The column was then eluted by the equilibrium buffer with gradient concentration of imidazole (50 mM, 100 mM and 300 mM). 12% SDS-PAGE evaluated the purification efficiency of protein. Protein complexes were purified by molecular-sieve chromatography (SEC) at 4 ℃ on Superdex-200 pre-grade column 16/100 (Pharmacia, GE Healthcare Bio-sciences, Pittsburgh, PA, USA), in 50 mM Tris-HCl buffer, pH 8, containing 150 mM NaCl, 8 M Urea (running buffer) at a flow rate of 1.0 mL min-1, and the elution profiles were continuously monitored with an online detector at a wave length of 280 and 260 nm. The target protein fractions were collected and protein purity was evaluated by SDS-PAGE.
After purification by affinity chromatography, the high purity of the target fraction was pooled and loaded in dialysis bag. The purified recombinant protein was renatured at 4 ℃ in buffer containing 1×PBS (Phosphate Buffered Saline, pH 7.4), 4 mM GSH, 0.4 mM GSSG, 2 mM EDTA, 0.4 mL Arginine, and 2 M Urea. After renaturation, fractions containing the protein were dialyzed against 1×PBS (pH 7.4) storage buffer for 6–8 h.
The dialyzates were separately loaded in tubes, freeze-dried, and then stored at –80 ℃; resulting protein powders were designated as the PhMEP1 fraction.
2.5. Immunoblot analysis
The separated proteins were electroblotted onto a polyvinylidene difluoride (PVDF) membrane, which was blocked with 5% non-fat milk in PBS buffer with 0.05% Tween 20 for 1 hour. The membrane was incubated overnight at 4℃ with Anti-His tag mouse monoclonal antibody (1:5000 dilution in blocking buffer), then washed, and incubated with HRP-conjugated goat anti-mouse antibody (1:5000). The proteins were detected using the chemiluminescence (CL) of luminol–H2O2 system.
2.6. Protease assay analysis
The enzyme activity was determined by the method of Ueda et al., 2013 with minor modification. The PhMEP1 activity was detected using azocasein. The reaction system is 1 mL. The mixture of 2% (w/v) azocasein solution, 350 μL of 0.1 M Tris-HCl buffer (pH 8.5) and 100 μL enzyme solution were incubated at 37 ℃ for 10 min. 500 μL of 10% trichloroacetic acid was then added to stop the reaction. The solution was left to stand at room temperature for 1 hour, then centrifuged with 12000×g for 10 min. One unit (U) of protease activity was defined as the amount of enzyme that will increase in absorbance of 0.001 at 280 nm min–1.
2.7. Inhibition experiment of metal ion chelating agent
The enzyme was incubated with an inhibitor (1,10-phenanthroline or EDTA) in 0.1 M Tris-HCl buffer (pH 8.5) at 25 ℃ for 1 hour. The inhibitors and their concentrations in the reaction system were as follows: EDTA (1 mM, 5 mM, 40 mM) and 1,10-phenanthroline (1 mM, 5 mM, 40 mM). The detection of the residual activity of PhMep1 after interaction with the inhibitors was based on the method of section above. The activity observed in the absence of inhibitor (control) was set to 100% (Ueda et al., 2013).
2.8. Subcelluar localization of PhMEP1
Solutions of Agribacterium tumefaciens-carrying pCambia1302-eGFP-PhMEP1 were infiltrated into leaf pavement cells of 4-week-old N. benthamiana plants. At two days post-infiltration (dpi), water-mounted slides of leaf tissue from agro-infected leaves were visualized by confocal microscopy. A. tumefaciens-carrying pCambia1302-eGFP was infiltrated into leaf cells as a control. Leaves were observed under a Nikon A1 confocal laser scanning microscopy.
3.1. Sequence analyses of PhMEP1 gene
The full-length cDNA sequence (3569 bp) of PhMEP1 was obtained through 2 round 3’-RACE PCRs from P. helianthi isolate SY and deposited in the GenBank database (GenBank accession no. KU994904). The whole sequence of protease was analyzed by ORF Finder, and a sequence of 2781bp was obtained. The ORF region of the whole protease sequence was further analyzed by Interpro. The predicted PhMEP1 contained a signal peptide sequence (at 1–22aa) and a M36 metalloprotease domain (at 483–928 aa). The M36 peptidase domain is responsible for catalytic activity and contains the active site motif HEXXH (at 697–702 aa) characteristic of Zn metalloproteases (Fig. 1A). Further secondary structure analysis revealed that PhMEP1 were composed of 27.14% alpha helix, 13.19% extended strand, 4.86% beta turn and 54.81% random coil, and random coil was major part (Fig. 1B). The predicted model of three-dimensional structure shows that PhMEP1 contained three residues are important Zn binding sites: His697, Glu727 and His788 (Fig. 1C). These results indicated that PhMEP1 should be a zinc metalloprotease in M36 family. A phylogenetic tree of MEPs from 20 representative species was constructed, revealing that PhMEP1 is most closely related to Puccinia graminis f. sp. Tritici (KAA1133194.1) (Fig. 1D).
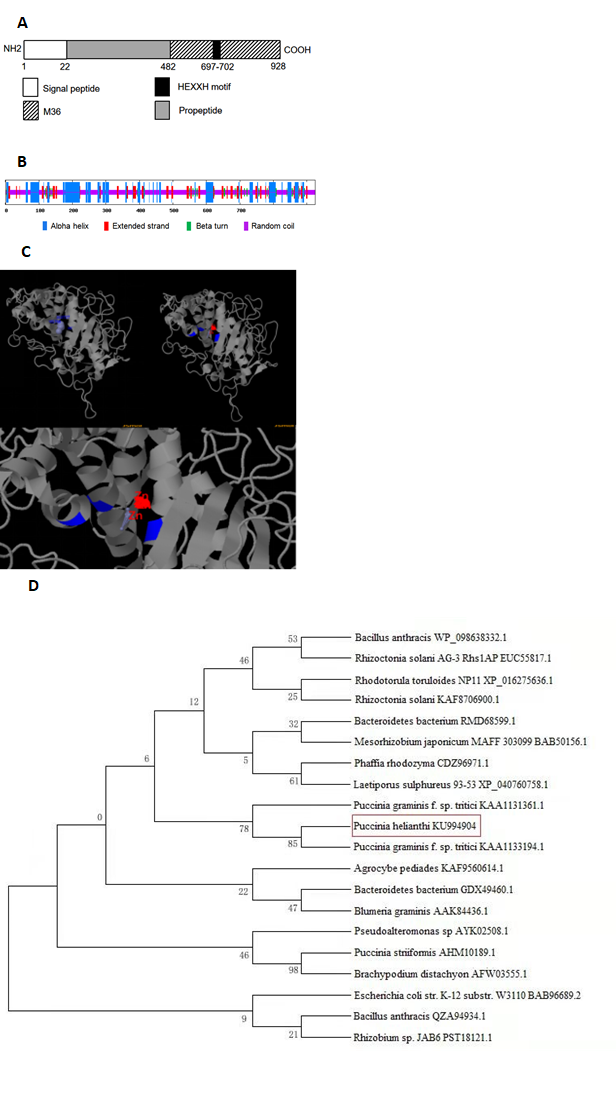
Fig. 1 Bioinformatic analysis of PhMEP1 protein. (A): The structural domain sites in PhMEP1 protein. (B): The secondary structure of PhMEP1. (C): The predicted model of PhMEP1 protein was made with the Phyre2 server. The residues implicate Zn binding sites are show in blue. (D): Phylogenetic analysis of 20 MEP proteins.
The full-length ORF sequence of PhMEP1 was amplified from the urediniospores of P. helianthi isolate SY. The newly formed vector was named pET30a- PhMEP1 and identified by colony PCR (Fig. 2A) and double-enzyme digestion with NdeI and HindIII (Fig. 2B).
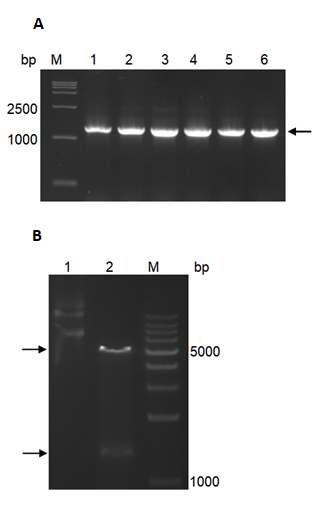
Fig. 2 Cloning and heterologous expression of PhMEP1. (A): Colony PCR of pET30a-PhMEP1 transformant in E. coli, using primers synthesized based on the amino acid sequences of the mature protein. Lanes: M, 15000 DNA Marker; 1–6, production of PCR. Target PCR product about 1300 bp is pointed with black arrowheads. (B): Restriction analysis of recombinant plasmid pET30a-PhMEP1. Two separate bands at about bp 5200 and 1300 could be seen. Lanes: M, DNA Marker; 1, plasmid DNA; 2, fragments generated from pET30a-PhMEP1 digested by NdeI and HindIII.
3.2. Expression and purification of PhMEP1 in E. coli
PhMEP1 was expressed successfully in E. coli strain BL21 (DE3) after induction of expression by IPTG for 16 h. The PhMEP1 fusion protein was obtained as expected (Fig. 3A).
The expressed fusion protein was harvested by ultrasonically lysing the E. coli. After centrifugation, bacterial supersonic lysates and culture supernatants were evaluated. Insoluble material from the lysates contained target protein (about 50 kDa). Supernatant obtained from the lysed E. coli cell contained some protein with similar molecular weight to the target protein and could not be adsorbed by Ni-IDA column and eluted after incubation with Ni-IDA. No target protein was found in eluting fractions after elution with 50 or 500 mM imidazole. Expressed fusion proteins existed in the form of inclusion body in E. coli by SDS-PAGE.
After dissolution of inclusion bodies in binding buffer containing 8 M Urea, the fusion protein was purified with metal affinity chromatography column. Eluted fractions containing target proteins were obtained with different concentration imidazole.
By size-exclusion chromatography (SEC), the protein/DNA complexes were eluted in a single peak and high purity protein was achieved. Fractions containing target protein were shown to be homogeneous by SDS-PAGE (Fig. 3B). Relatively high purity protein was pooled to be dialysed, and eventually concentrated to a final protein concentration of 0.6 mg mL-1 (before freeze drying).
Purified protein followed by SDS-PAGE revealed a band at about 50 kDa. This 50 kDa band represented the P. helianthi PhMEP1 further showed strong immunological cross-reactivity with anti-His mouse monoclonal antibody prepared against the His fusion protein of P. helianthi PhMEP1 (Fig. 3C).
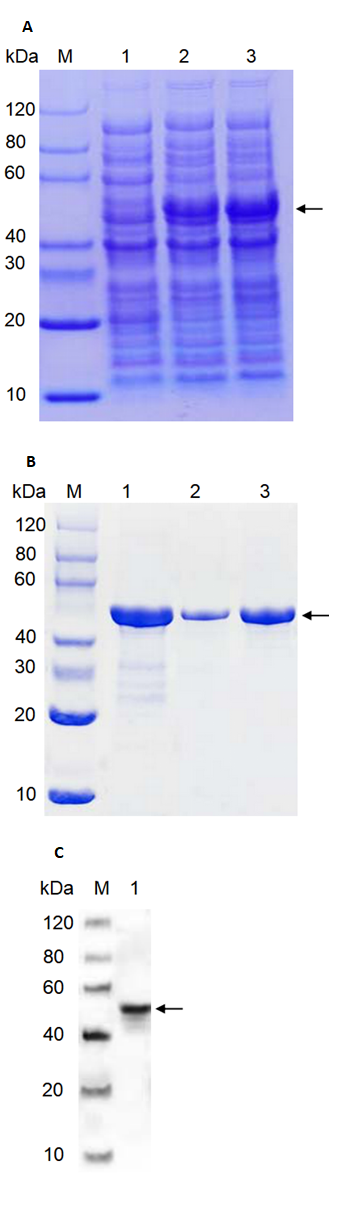
Fig. 3 Expression and purification of PhMEP1. (A): SDS-PAGE of cell lysates of E. coli transformants containing the proteinase expression vector. Lanes: M, SDS-PAGE protein marker; 1, uninduced control; 2, induced with IPTG for 16 h at 15 ℃; 3, induced with IPTG for 16 h at 37 ℃. Target protein about 50 kDa is pointed with black arrowheads (B): SDS-PAGE analysis of purified proteins by SEC. Lanes: M, protein marker; 1, purified sample; 2,3, fractions collected after purified by SEC. Target protein about 50 kDa is pointed with black arrowheads. (C): Western blot analysis. Western hybridization of 2 μg His fusion protein with anti-His mouse monoclonal antibody in 1:5000 dilution M, Western blot marker. 1, PhMEP1 protein (2.0 μg) ; Target protein is about 50 kDa pointed with black arrowheads.
3.3. Protease activity of PhMEP1
The results of azocasein hydrolysis showed that PhMEP1 could hydrolyze azocasein substrate. The specific activity was 1560 U mg–1.
PhMEP1 activity was suppressed by 1,10-phenanthroline and EDTA. The activity of PhMEP1 was decreased step by step with the increasing inhibitor concentration. When the concentration of 1,10-phenanthroline and EDTA is 1 mM, the activity of purified PhMEP1 was reduced by 49% and 40% respectively. When the concentration of 1,10-phenanthroline and EDTA is 5 mM, the activity of purified PhMEP1 was reduced by 90% and 80% respectively. When the concentration of 1,10-phenanthroline and EDTA is 40 mM, the activity of purified PhMEP1 was reduced by 99% and 96% respectively (Table 1).
Table 1. Effect of protease inhibitors on enzyme activity
|
Inhibitors |
Concentration (mM) |
Relative activity (%) |
|
Control |
|
100 |
|
1,10-phenanthroline |
1 |
51 |
|
5 |
10 |
|
|
40 |
1 |
|
|
EDTA |
1 |
60 |
|
5 |
20 |
|
|
40 |
4 |
3.4. The subcellular localization of PhMEP1
To determine the subcellular location of PhMEP1, the full coding sequence of PhMEP1without the stop codon was subcloned in fusion to the N-terminus of GFP, and the pCambia1302-eGFP-PhMEP1 fusion-expressing vector was generated. The resulting pCambia1302-eGFP-PhMEP1 and the control pCambia1302-eGFP were individually introduced into N. benthamiana leaf sell. These fluorescent proteins were transiently expressed and observed via a confocal laser scanning microscopy. The fluorescent images in N. benthamiana leaf sell showed that PhMEP1-GFP might be distributed at the plasma membrane, but not in chloroplast, cell nucleus and cytoplasm, whereas GFP protein alone was distributed in the plasma membrane and nucleus (Fig. 4).
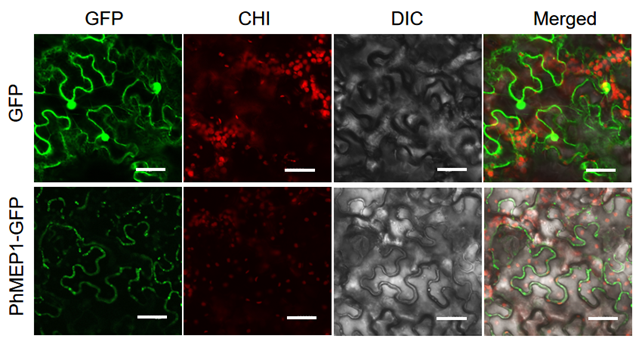
Fig. 4 Subcellular localization of the PhMEP1. The mature coding sequences of PhMEP1 was cloned in frame with GFP. Live cell imaging using confocal microscope of epidermal cells of 2-days-old N. benthamiana transient expression PhMEP1-GFP plantlets. Bar=20 µm
In this research, we performed prokaryotic expression and described the characteristics of a fungalysin-like metalloprotease from P. helianthi. This is the first time a fungalysin-like metalloprotease has been identified from P. helianthi. Little is known about the function, specificity, and modulatory mechanism of the extracellular proteases of fungus, so it is difficult to speculate the roles of the individual M36 fungal proteases in P. helianthi. However, the extensive understanding of this gene family provides a good model system for learning the roles of the similar individual peptidases in the extracellular condition.
Extracellular proteases play a vital role as hydrolases that maintain and adjust the nutritional requirements of microorganisms and are widely accepted in the food and pharmaceutical industries (Adekoya et al., 2009). The pathogen's extracellular proteases are essential for regular growth and in favor of multiple ways of bacterial infection. Among the secreted proteases, elastase and alkaline protease of Pseudomonas aeruginosa are the two most recognized and understood metalloproteases (Matsumoto 2004). Structurally, Mep active centers contain one or two metal ions, most of which are zinc-containing proteins. In fact, many zinc-containing bacterial proteases and fungal proteases are found throughout the world within microorganisms Chen et al., 2015; Ishii et al., 2014; Wu et al., 2011; Li et al., 2014), such as the AprX extracellular metalloproteinases of many fluorescent pseudomonads (Dufour et al., 2008; Zhang et al., 2009), members of the Bacillus neutral protease family (Wu et al., 2011) the SmP metalloprotease (serralysin) of Serratia marcescens (Chen et al., 2015), the RpA metalloprotease (serralysin) of Ralstonia pickettii and alkaline protease of fungi. Extracellular Meps are known to be important virulence factors related to bacterial and fungal pathogenicity (Zhang et al., 2017). Because of the great medical, physiological and commercial potential of these enzymes in practical applications, it is essential to isolate and screen various microbial strains.
Along with the results of the bioinformatic, the activity of P. helianthi PhMEP1 that was suppressed by 1,10-phenanthroline and EDTA strongly suggested that the product of the cloned gene was a metalloprotease. The localization of PhMEP1 in planta can help us investigate if PhMEP1 associates with host molecules for interaction.
Taken together, our results suggest that PhMEP1 manipulates plants molecules by targeting plasma membrane of epithelial cells, and degrading cellular constituent via protein-binding, which is a pathogenicity factor contributing to penetration and infection by rust pathogen.
P. helianthi is an obligate pathogen of sunflower. The enzyme PhMEP1 secreted by this pathogen would assist the invading fungus to penetrate through host barriers and thus adding pathogen virulence. Since the use of extracellular enzymes to degrade the structural barriers in the host appears to be a commonly used strategy of fungal pathogens (Mercer et al., 2019; Podder et al., 2019), therapeutic approaches targeting against the major virulence factors would be a highly desirable alternative to prevent and treat sunflower rust.
We reported the characterization of an extracellular PhMEP1 gene from P. helianthi, which is a causal agent of sunflower rust. Its molecular mass was about 50 kDa. The PhMEP1 activity could be suppressed by 1,10-phenanthroline and EDTA. It could interact with the cell wall, or the plasma membrane of host plant. It is a pathogenicity factor contributing to penetration and infection by rust pathogen.
The authors declare that they have no known competing financial interests or personal relationships. This study was funded by the National Natural Science Foundation of China (No. 32060598, 31760509), the Natural Science Foundation of Inner Mongolia Autonomous Region (No. 2020MS03046).
Silva, R., Cabral, T., Rodrigues, A., 2013. Production and partial characterization of serine and metallo peptidases secreted by Aspergillus fumigatus Fresenius in submerged and solid-state fermentation. BRAZ J MICROBIOL. 44(1): 235-243. DOI: 10.1590/s1517-83822013000100034 PMid:24159310
View Article PubMed/NCBISharpton, T.J., Stajich, J.E., Rounsley, S.D. et al., 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19: 1722-1731. DOI: 10.1101/gr.087551.108 PMid:19717792
View Article PubMed/NCBISanz-Martín, J.M., Pacheco-Arjona, J.R., Bello-Rico, V., Vargas, W.A., Monod, M., Díaz-Mínguez, J.M., Sukno, S.A., 2016. A highly conserved metalloprotease effector enhances virulence in the maize anthracnose fungus Colletotrichum graminicola. Mol Plant Pathol. 17, 1048-1062. DOI: 10.1111/mpp.12347 PMid:26619206
View Article PubMed/NCBIPodder, S., Saha, D., Ghosh, T.C., 2019. Deciphering the intrinsic properties of fungal proteases in optimizing phytopathogenic interaction. Gene. 711, 143934. DOI: 10.1016/j.gene.2019.06.024 PMid:31228540
View Article PubMed/NCBINaumann, T.A., Wicklow, D.T., Price, N.P., 2011. Identification of a chitinase-modifying protein from Fusarium verticillioides truncation of a host resistance protein by a fungalysin metalloprotease. J Biol Chem. 286, 35358-35366. DOI: 10.1074/jbc.M111.279646 PMid:21878653
View Article PubMed/NCBIMonod, M., Paris, S., Sanglard, D., Jaton-ogay, K., Bille, J., Latge, J.P., 1993. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect Immun. 61, 4099-4104. DOI: 10.1128/IAI.61.10.4099-4104.1993 PMid:8406798
View Article PubMed/NCBIMatsumoto, K., 2004. Role of bacterial proteases in pseudomonal and serratial keratitis. Biol Chem. 385, 1007-1016. DOI: 10.1515/bc.2004.131 PMid:15576320
View Article PubMed/NCBIMercer, D.K., Stewart, C.S., 2019. Keratin hydrolysis by dermatophytes. Med Mycol. 57, 13-22. PMid:29361043
View Article PubMed/NCBIMarkell, S.; Gulya, T.J.; McKay, K.; Hutter, M.; Hollingsworth, C.; Ulstad, V.; Koch, R.; Knudsvig, A., 2009. Widespread occurrence of the aecial stage of sunflower rust caused by Puccinia helianthi in North Dakota and Minnesota in 2008. Plant Dis. 93, 668-668. DOI: 10.1094/PDIS-93-6-0668C PMid:30764420
View Article PubMed/NCBILilly, W.W., Stajich, J.E., Pukkila, P.J., Wilke, S.A., Inoguchi, N., Gathman, A.C., 2008. An expanded family of fungalysin extracellular metalloproteases of Coprinus cinerea. Mycol. Res. 112, 389-398. DOI: 10.1016/j.mycres.2007.11.013 PMid:18313909
View Article PubMed/NCBILi, X., Song, Y., Lu, Y., Jing, L., 2019. Cloning and bioinformatics analysis of elastinolytic metalloproteinase gene from Puccinia helianthi Schw. Acta. Agriculturae Boreali-Sinica. 34, 186-193. DOI: CNKI:SUN:HBNB.0.2019-03-031
Li, J., Zhang, K.Q., 2014. Independent expansion of zincin metalloproteinases in Onygenales fungi may be associated with their pathogenicity. PLoS. ONE. 9(2), e90225. DOI: 10.1371/journal.pone.0090225 PMid:24587291
View Article PubMed/NCBIKim, J.S., Kim,J.E., Choi,B.S. et al. 2008. Purification and characterization of fibrinolytic metalloprotease from Perenniporia fraxinea mycelia. Mycol. Res. 112, 990-998 . PMid:18550350
View Article PubMed/NCBIJousson, O., Léchenne, B., Bostems, O., Capoccia, S., Mignon, B., Barblan, J., Quadroni, M., Monod, M., 2004. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology. 150, 301-310. DOI: 10.1099/mic.0.26690-0 PMid:14766908
View Article PubMed/NCBIJing, L., Guo, D., Hu, W.J., Niu X., 2017. The prediction of a pathogenesis-related secretome of Puccinia helianthi through high-throughput transcriptome analysis. BMC Bioinformatics. 18, 166. DOI: 10.1186/s12859-017-1577-0 PMid:28284182
View Article PubMed/NCBIJia, Y., McAdams, S.A., Bryan, G.T., Hershey, H.P., Valent, B., 2000. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. The EMBO Journal. 19, 4004-4014. DOI: 10.1093/emboj/19.15.4004 PMid:10921881
View Article PubMed/NCBIIshii,K., Adachi, T., Hamamoto, H., Sekimizu, K., 2014. Serratia marcescens suppresses host cellular immunity via the production of an adhesion-inhibitory factor against immunosurveillance cells. J. Biol. Chem. 289, 5876-5888. DOI: 10.1074/jbc.m113.544536 PMid:24398686
View Article PubMed/NCBIGulya, T.J.; Harveson, R.; Mathew, F; Block, C.C.; Thompson, S.; Kandel, H.; Berglund, D.; Sandbakken, J.; Kleingartner, L.; Markell, S., 2019. Comprehensive disease survey of U.S. sunflower: disease trends, research priorities and unanticipated impacts. Plant Dis. 103, 601-618. DOI: 10.1094/PDIS-06-18-0980-FE PMid:30789318
View Article PubMed/NCBIDufour, D., Nicodeme, M., Perrin, C., Driou, A., Brusseaux, E., Humbert, G., Gaillard, J.L., Dary, A., 2008. Molecular typing of industrial strains of Pseudomonas spp. isolated from milk and genetical and biochemical characterization of an extracellular protease produced by one of them. Int. J. Food Microbiol. 125, 188-196. DOI: 10.1016/j.ijfoodmicro.2008.04.004 PMid:18511140
View Article PubMed/NCBIChen, C.M., Liu, J.J., Chou, C.W., Lai, C.H., Wu, L.T., 2015. RpA, an extracellular protease similar to the metalloprotease of serralysin family, is required for pathogenicity of Ralstonia pickettii. J. Appl. Microbiol. 119, 1101-1111. DOI: 10.1111/jam.12903 PMid:26184602
View Article PubMed/NCBIAdekoya, O.A., Sylte, I., 2009. The thermolysin family (M4) of enzymes: therapeutic and biotechnological potential. Chem. Biol. Drug Des. 73, 7-16. DOI: 10.1111/j.1747-0285.2008.00757.x PMid:19152630
View Article PubMed/NCBITallant, C., García-Castellanos, R., Seco, J., Baumann, U., Gomis-Rüth, F.X., 2006. Molecular analysis of ulilysin, the structural prototype of a new family of metzincin metalloproteases. J Biol Chem. 281, 17920-17928. DOI: 10.1074/jbc.M600907200 PMid:16627477
View Article PubMed/NCBIUeda, M.,Yamamoto, N., Kusuda, M., Nakazawa, M., Takenaka, S., Miyatake, K., Ouchi, K., Sakaguchi, M., Inouye, K., 2013. Purification and characterization of a new fungalysin-like metallopeptidase from the culture filtrate of a plant worm, Nomuraea atypicola. Process Biochem. 48, 190-194. DOI: 10.1016/j.procbio.2012.11.005
View ArticleWu, J.W., Chen, X.L., 2011. Extracellular metalloproteases from bacteria. Appl Microbiol Biotechnol. 92, 253-262. DOI: 10.1007/s00253-011-3532-8 PMid:21845384
View Article PubMed/NCBIZhang, W.W., Hu, Y.H., Wang, H.L., Sun, L., 2009. Identification and characterization of a virulence-associated protease from a pathogenic Pseudomonas fluorescens strain. Vet Microbiol. 139(1-2), 183-188. DOI: 10.1016/j.vetmic.2009.04.026 PMid:19464828
View Article PubMed/NCBIZhang, X., Wang, Y., Chi, W., Shi, Y., Chen, S., Lin, D., Jin, Y., 2014. Metalloprotease genes of Trichophyton mentagrophytes are important for pathogenicity. Med Mycol. 52, 36-45. DOI: 10.3109/13693786.2013.811552 PMid:23859078
View Article PubMed/NCBIZhang, Y., Yan, J., Ge, Y., 2017. Advances in bacterial extracellular metalloproteases and their pathogenic roles. Chin J Microbiol Immunol. 37161-37164. DOI: 10.3760/cma.j.issn.0254-5101.2017.02.013
Zhou, R., Zhou, X., Fan, A., Wang, Z., Huang, B., 2018. Differential functions of two metalloproteases, Mrmep1 and Mrmep2, in growth, sporulation, cell wall integrity, and virulence in the filamentous fungus Metarhizium robertsii. Front Microbiol. 9, 1528. DOI: 10.3389/fmicb.2018.01528 PMid:30034386
View Article PubMed/NCBI