Priyanka Kundu
Email: priyankakundu.72@gmail.com
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 2
Page No: 376-391
Priyanka Kundu
Email: priyankakundu.72@gmail.com
Priyanka Kundu1*, Vinay Kumar Singh2 ( vinaysingh@bhu.ac.in)
1School of Biotechnology, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India PIN-221005
2Centre for Bioinformatics, School of Biotechnology, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India PIN-221005
Ines Mancini(ines.mancini@unitn.it)
Md Tabish Rehman(m.tabish.rehman@gmail.com)
Hai Qian(qianhai24@163.com)
Priyanka Kundu, Vinay Kumar Singh, Molecular docking of Ibuprofen and its derivatives with novel cancer targets: Implication of anticancer therapy for novel targets (2020) Journal of Computational Chemistry & Molecular Modeling 4(2) pp:376-391
Epidemiological and clinical studies suggest that the nonsteroidal anti-inflammatory drug (NSAID)-Ibuprofen’s anti-carcinogenic property is attributable due to its capacity to inhibit the cyclooxygenase (COX) enzyme (both the isoforms-COX1 and COX-2), inhibit proliferation and induce apoptotic cell death. However, the precise molecular mechanism for the anti-cancer activity of Ibuprofen is still not fully understood. As the COX-dependent mechanisms were already established now the COX-independent mechanisms are targeted along with newly synthesized Ibuprofen derivatives to develop safer and more efficacious drugs for cancer chemo-prevention. Here, the interactions of Ibuprofen and its derivatives (Carboxy-Ibuprofen, Hydroxy-Ibuprofen and Methyl ester Ibuprofen) with LDH-A, Survivin, Glucocorticoid Receptor and Androgen receptor were analysed by PatchDock and YASARA (Yet Another Scientific Artificial Reality Application).The docking ability of the drug with the targets were based on the analysis of dissociation constant (Kd), geometric shape complementary score (GSC score), approximate interference area (AI area) and binding energy along with analysis of drug likeness parameters of Ibuprofen and its derivatives. The outcome of this study sheds light on the efficacy drug derivatives and identifying novel target molecule for cancer chemoprevention which can be further used for in vivo studies.
Keywords: Ibuprofen, NSAIDs, In-silico, Drug derivatives, Molecular docking, Drug likeness, Molecular targets, Cancer therapeutics;
Ibuprofen is the nonsteroidal anti-inflammatory drug (NSAID) class that is used for treating pain, fever and inflammation. Prior epidemiological and clinical studies have suggested effectiveness of long-term and regular use of Ibuprofen in treatment and risk reduction for 7-10 malignancies along with the four major types of cancer: colon, breast, lung, and prostate (Harris et al.,2005;Thun et al.,2002;Janssen et al.,2006;Bittoni et al.,2017).The anti-carcinogenic property is attributable due to its capacity to inhibit the cyclooxygenase (COX) enzyme(both the isoforms-COX1 and COX-2), inhibit proliferation and induce apoptotic cell death (Giardiello et al.,1995; Thun et al.,2002; Leidgens et al.,2015). However, the chemopreventive efficacy may also be exerted due to their COX-independent mechanism of action where they act on the non COX targets, showing different cellular effects (Gurpinar et al., 2013). Novel Ibuprofen derivatives can be synthesized by different modifications of Ibuprofen that would have better/similar significant effect and lesser side effects than the parent Ibuprofen in their chemopreventive efficacy (Ouyang et al., 2013). In silico molecular docking with physicochemical properties and drug likeness parameters can be used for computer-aided drug designing and the newly synthesized Ibuprofen derivatives. These can be used in model in vivo studies, for identifying and characterizing alternate therapeutic (molecular) targets and elucidation of additional biochemical process leading to the development of safer and more efficacious drugs for cancer chemoprevention, also saving valuable time, money and resources.
The COX-dependent mechanism is well established so now in this study, COX- independent mechanisms are being targeted along with newly synthesized Ibuprofen derivatives to develop safer and more efficacious drugs for cancer chemo-prevention. Here, Ibuprofen(IBP) (2-(4-Isobutylphenyl) propanoic acid) and its derivatives - Carboxy-IBP, Hydroxy-IBP(2-Hydroxy-Ibuprofen) and IBP methyl ester are considered for its chemopreventive effect on the COX- independent targets-LDH-A, Survivin, Glucocorticoid Receptor (GR) and Androgen Receptor (AR). As metabolites of IBP, Carboxy-IBP is dicarboxylic acid, i.e. IBP in which one of the methyl groups in the isobutyl portion has been converted to the corresponding carboxylic acid, 2-Hydroxyibuprofen is a hydroxy monocarboxylic acid, i.e. IBP in which the methine proton on the isobutyl group has been replaced by a hydroxy group and Methyl 2-(4-isobutylphenyl) i.e. IBP methyl ester propanoate is the methyl ester of IBP.
Lactate dehydrogenase A (LDH-A) executes the final step of aerobic glycolysis that has been reported to be involved in the tumor progression. Studies show abnormal expression of LDHA has been observed in many human cancers, such as pancreatic cancer (Shi et al., 2014), hepatocellular carcinoma (Sheng et al., 2012), breast cancer (Zhao et al., 2009) and prostate cancer (Xian et al., 2015). Inhibition of LDHA reduced cell malignant transformation and remarkably delayed tumor formation, indicating that the underlying role of LDH-A in tumor initiation or maintenance, thus revealing the oncogenic role of LDHA in prostate cancer which also suggest that LDHA might be a potential therapeutic target in anticancer therapy (Di Stefano et al., 2016).
Survivin is a member of the inhibitor of apoptosis protein (IAP) that blocks apoptotic pathways by inhibition of caspase and pro-caspase molecules (Ambrosini et al.,1997; Tamm et al., 1998) .It is a tumor antigen and overexpressed in human cancers, giving rise to peptides eliciting spontaneous CD8+ and CD4+ responses. Survivin has the dual function of blockade of apoptosis and regulation of cell division (Friedrichs et al., 2006; Dohi et al., 2004; Margulis et al., 2007). IBP treat shows a gradual decrease in expression of surviving in a time-dependent manner, thus due to its direct association with tumor survival, it is regarded as an ideal target structure for anti-cancer therapy (Greenspan et al., 2011).
Various studies show that Glucocorticoid Receptor (GR) functional activity appears to be highly context-dependent (Conzen S. D., 2017). GR is a predominately cytoplasmic protein whose ligand-binding conformation is maintained by its chaperoning partners in a large multi- protein complex. It is able to shuttle between the nucleus and the cytoplasm and can help or inhibit the expression of specific genes via different mechanisms- transactivation, which promotes the transcription of glucocorticoid responsive element (GRE)-driven genes (e.g. GILZ), and trans repression, via which GR inhibits the expression of genes mediated by transcription factors such as NF-κB and AP-1(Hache et al.,1999; Vandevyver et al.,2012; Sundahl et al.,2016; Ratmanet al.,2013; Clarisseet al.,2018).With detailed knowledge of GR signaling it has been a target for prostate, breast and various other cancers for therapy (Bakker et al.,2017; Kachet al.,2015).
Androgen receptor (AR) is a steroid receptor transcriptional factor for testosterone and dihydro-testosterone, which consists of four main domains, the N-terminal domain, DNA-binding domain, hinge region, and ligand-binding domain (Fujita et al., 2019). AR plays pivotal roles in prostate cancer and prostate cancer therapy. The inhibition of AR activity through mechanisms in addition to androgen ablation, such as modulation of signal transduction pathways, may delay prostate cancer progression thus making it a potent target for therapy (Miyamoto et al., 2008; Huret al., 2004).
Based on proper review literature the 3D structures of the target receptors and ligands were retrieved from Protein Data Bank (https://www.rcsb.org/) and PubChem (https://pubchem.ncbi.nlm.nih.gov/) databases, respectively. The 3D structure of the target proteins was validated thorough BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) , RAMPAGE (http://mordred.bioc.cam.ac.uk/~rapper/rampage.php) , ResProx (http://www.resprox.ca/) , ERRAT (https://servicesn.mbi.ucla.edu/ERRAT/) and PDBSum (http://www.ebi.ac.uk/thornton-srv/databases/cgi-bin/pdbsum/) server and VADAR 1.8 (http://vadar.wishartlab.com/). The Active binding sites were identified by MetaPocket (https://projects.biotec.tu-dresden.de/metapocket/) server. Docking was performed by PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) server and YASARA (http://www.yasara.org/macros.htm) tool, whereas docking complexes were visualized and edited by Discovery Studio 3.5 (http://accelrys.com/products/collaborative-science/biovia- discovery- studio/visualization- download.php) . The druglikeness was analyzed through Lipinski (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) filter, SwissADME (http://www.swissadme.ch/), admetSAR (http://lmmd.ecust.edu.cn/admetsar1/predict/) and FAF-Drugs4 (http://fafdrugs4.mti.univ-paris-diderot.fr/) . The methodology is depicted in a flow chart in Fig 1.
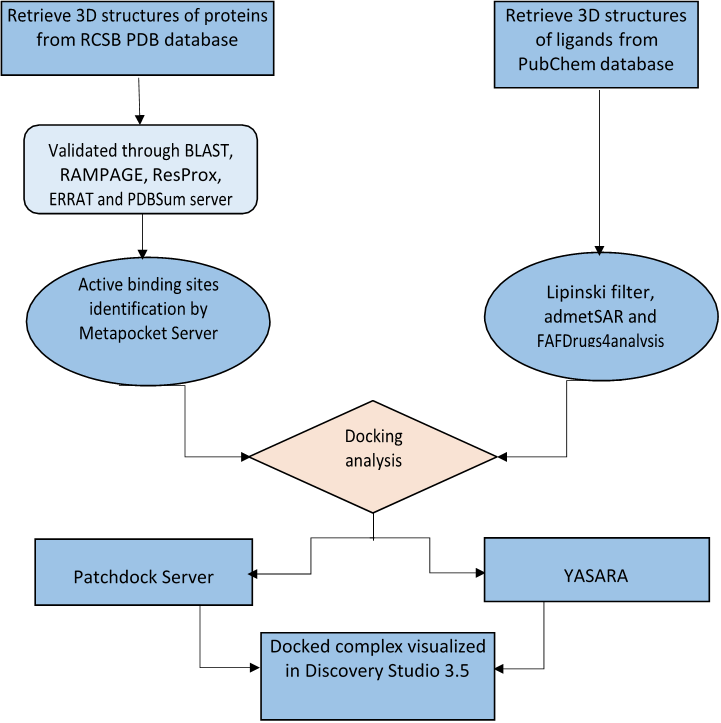
Fig 1. Flow chart depicting schematic methodology of in silico analysis.
2.1.Retrieval of target receptor structures
Protein Data Bank (https://www.rcsb.org/) was used for retrieving the structure of target receptors LDH-A (PDB:4ZVV, DOI:10.2210/pdb4ZVV/pdb), Survivin (PDB:3UEC, DOI: 10.2210/pdb3UEC/pdb), Glucocorticoid Receptor (GR) (PDB:4UDD, DOI:10.2210/pdb4UDD/pdb) and Androgen Receptor (AR) (PDB:3L3X, DOI:10.2210/pdb3L3X/pdb) in cancer in Homo sapiens, which are recognized as targets of Ibuprofen and its derivatives- Carboxy-Ibuprofen, Hydroxy- Ibuprofen and Ibuprofen methyl ester. The criteria for selection of the structures were based on PDB advance BLAST, RAMPAGE, ResProx, ERRAT and PDBsum analysis. The structures used in this study were those displaying the maximum score and query cover in BLAST as the first criteria of selection.
2.2. Retrieval of ligand structures
The structures of Ibuprofen (CID: 3672, https://pubchem.ncbi.nlm.nih.gov/compound/3672) and its derivatives-Carboxy-Ibuprofen (CID: 10444113, https://pubchem.ncbi.nlm.nih.gov/compound/10444113), 2-Hydroxy-Ibuprofen (Hydroxy-IBP) (CID: 10443535,https://pubchem.ncbi.nlm.nih.gov/compound/10443535) and Ibuprofen methyl ester (CID: 109101,https://pubchem.ncbi.nlm.nih.gov/compound/109101) were retrieved from the PubChem (https://pubchem.ncbi.nlm.nih.gov/) database. These structures were further used for docking calculation. The selected 3D structure of the ligands was retrieved from PubChem Compound database in SDF format followed by conversion in the PDB format and optimization using Discovery Studio 3.5 visualizer.
2.3.Active binding site prediction
Prior to the docking analysis, the active binding site prediction of LDH- A (PDB:4ZVV), Survivin (PDB:3UEC), Glucocorticoid Receptor (GR) (PDB:4UDD) and Androgen Receptor (PDB:3L3X) were carried out by MetaPocket 2.0 (https://projects.biotec.tu-dresden.de/metapocket/) server, retrieving top 3 major binding pockets for analysis of active binding residues and comparison of the docking results.
2.4.Docking analysis
PatchDock (https://bioinfo3d.cs.tau.ac.il/PatchDock/php.php) server, a geometry based molecular docking algorithm, was used for docking analysis of IBP and its derivatives to target receptors. The PDB files of the ligand and receptors were uploaded to PatchDock server for docking analysis, keeping the cluster RMSD at default value of 4.0 and protein- small ligand complex type as the analysis parameters. Analysis on PatchDock server yielded results for the geometric shape complementarity score (GSC score) and approximate interface area (AI area). Additional docking tool YASARA (http://www.yasara.org/macros.htm) (Yet Another Scientific Artificial Reality Application), an AutoDock based tool for molecular docking and virtual screening was used for analyzing dissociation constant (Kd) and binding energy of the docked complexes. The more binding energy of a ligand during a molecular dynamic simulation indicates a better binding (Yadav et al., 2017).
2.5.Analysis of drug likeness of IBP, Carboxy-IBP, Hydroxy-IBP and IBP methyl ester
The drug likeness prediction of IBP, Carboxy-IBP, Hydroxy-IBP and IBP methyl ester was carried out by the Lipinski filter (http://www.scfbio-iitd.res.in/software/drugdesign/lipinski.jsp) , according to the Lipinski Rule of 5 , an orally active drug should comply to a minimum of four of the five laid down criteria for druglikeness namely: molecular mass, cLogP, hydrogen donor and acceptor and molar refractive index . SwissADME (http://www.swissadme.ch/) computed the physicochemical descriptor, predicted the ADME properties, pharmacokinetic parameters, druglike nature and medicinal chemistry friendliness. Along with that admetSAR(http://lmmd.ecust.edu.cn/admetsar1/predict/) was used to analyze the properties of ligand with respect to prediction of adsorption, distribution, metabolism, excretion and toxicity (ADMET), which is reported as an useful tool in drug discovery. This tool was utilized for predicting important descriptors of drug likeness. FAFDrugs4(http://fafdrugs4.mti.univ-paris-diderot.fr/) was used to predict additional ADMET properties of IBP and its derivatives, which assist in filtering studies for selection of good drug candidates for drug development projects. The SDF (Structure Data Format) file of the IBP and its derivatives were downloaded from PubChem Database to calculate ADME properties using default parameters.
3.1.Retrieval of the 3-D structure of the target receptors and ligands
The structural details of target receptors LDH-A (PDB: 4ZVV), Survivin (PDB: 3UEC), Glucocorticoid Receptor (PDB: 4UDD) and Androgen Receptor (PDB: 3L3X) were obtained from the PDB data bank are presented in Fig 2.a)-2.d). The structures of Ibuprofen (CID: 3672) and its derivatives- Carboxy-Ibuprofen (CID: 10444113), 2-Hydroxy-Ibuprofen (Hydroxy-IBP) (CID: 10443535) and Ibuprofen methyl ester (CID: 109101) were retrieved from the PubChem database as presented in Fig. 3.a)-3.d).
Fig. 2.a)-d) 3D structures of retrieved receptors. Structures of LDH-A, Survivin, Glucocorticoid Receptor and Androgen Receptor retrieved from the PDB data bank.
Fig. 3.a)-d) 2D structures and 3D Conformers of IBP and its derivatives-Carboxy-IBP, Hydroxy-IBP and IBP methyl ester from the PubChem compound database.
3.2. Docking analysis of IBP and its derivatives
The results of the docking analysis with PatchDock server and YASARA, which are reported to be highly used and reliable tools for molecular docking analysis are presented in Fig. 4.a)-4.e) and the docking calculation depicting the ligand name, receptor name, reported active site, predicted/prominent active sites from MetaPocket 2.0 up to 3 number of pockets were determined, contacting receptor residues from YASARA and their common residues are shown in Table 1.
Table 1. Docking calculations depicting the ligand name, receptor name, reported active site, contacting receptor residues and their common residues.
|
Ligand Name |
Receptor Name |
Reported Active Site Residues |
Predicted Active Site |
Contacting receptor residues |
Common Residues |
|
IBUPROFEN (CID: 3672 ) |
LDH-A (PDB ID:4ZVV) |
Gly28,Ala29,Val30,Asp51, Val52,Ile53,Thr94,Ala95, Gly96,Arg98,Ile115,Val135, Asn137,Trp147,Pro153, Lys154,Leu164,Asp165, Arg168,Arg170,His185, His192,Gly193,Asp194, Ala237,Tyr238,Ile241, Gly245,Thr247,Ile251 |
Binding Site Id: 1 Ser160,Gly161,Leu164,His192,Ile251,Val135, Val139,Asp165, Val25,Gly26,Val50,Asp51,Val52, Tyr82,Ile119,Lys80,Val30, Asn137,Ser136,Ala95, Phe118,Ile115,Thr247,Thr94,Arg168,Val27 , Gly28,Ala29,Gly31,Ser248,Gly96,Gly193, Val233,Ser196,Pro138 ,Asp140,Glu191,Leu322 , Ala237,Val234,Asp194,Ile141,Asn114, Ile53,Lys56 , Tyr246,Ser236,Asn112,Ilea241,Ala97,Ser195,Thr321, Arg98,Val240 ,Tyr238 ,Ile325,Gly245 ,Leu108,Val109, Arg105, Gln99,Glu239,Ser104,Asn107,Lys242, Glu103,Gln100,Ser318, Leu106,Glu328,Gly324,Asp320 Binding Site Id: 2 Trp249,Ala250,Leu253,Ala167,Leu164,Arg168,Ser236,Ala237, Val240,Tyr171,Leu172,Lys41,Asp257,Glu260,Lys244, Thr247, Ser248,Arg170,Pro181,Leu182,Asn163,Val269 , His270 ,Pro271, Trp187 ,Ser254,Ser166,Gly186,His185 , His180,Arg268,Asp165, Ile251,Val233,Gln232,Glu239,Glu235 |
Arg168,Tyr171,Leu172, Ser236,Val240, Lys244, Thr247 ,Ser248,Trp249, Ala250,Ile251 |
Binding Site Id: 1 Arg168,Thr247,Ile251
Binding Site Id: 2 Arg168,Thr247,Ile251 |
|
SURVIVIN (PDB ID:3UEC) |
Phe13,Leu14 ,Lys15,Asp16, Arg18,Gly30,Glu40,Glu51, Leu54,Cys57,Cys60,Lys62, Glu63,Leu64,Glu65,Gly66, Trp67,Asp70,Asp71,Glu76,His77,His80,Cys84,Phe86, Val89,Gln92,Glu94,Glu95,Thr97,Gly99,Glu107,Lys120, Glu123,Ala128,Val131,Arg132,Ala139,Asp142 |
Binding Site Id: 1 Phe13,Phe86,Val89,Phe93,Leu96,Leu104,Phe59, Lys91,Arg18,Leu14,Gln92,Glu94,Lys90,Lys15,Leu87, Ser88,Glu40,Ala85,Ala41,Glu95,Lys78,Asp16,Ile74,His17,Ile19 Binding Site Id: 2 Lys62,Lys115,Asn118,Lys122,Cys60,Ala114,Asn111, Ser81,Gln56,Phe61,Glu63,Asn119,His80,Leu64 ,Leu54,Glu51,Glu65 |
Leu6,Trp10,Phe93, Glu94, Glu95,Leu96,Thr97,Leu98,Phe101 |
Binding Site Id: 1 Glu94,Glu95
Binding Site Id: 2 No Match
|
|
|
GLUCOCORTICOIDS (PDB ID:4UDD) |
Glu540,Pro541,Met560, Leu563,Asn564,Gly567, Gln570,Trp577,Met604, Trp610,Arg611,Tyr640, Asp641, Gln642,His645, Met646,Tyr648,Tyr660, Lys667,Met691,Lys695 Val702,Ser708,Trp712, Phe715,Glu727,Asn731
|
Binding Site Id: 1 Asn564,Trp600,Met601,Cys736,Phe749,Leu753,Leu563, Gly567,Thr739,Leu732,Tyr735,Met604,Gln570,Phe623, Met560,Ala605, Met646,Ile747,Gln642,Leu608,Leu566,Arg611, Cys643,Met639,Ile559,Ile629,Asp641,Asn731,Lys644, His645,Asn734,Gln738,Glu730,Met745,Thr556,Leu733, Thr562,Met565,Leu603,Ala607,Trp557 Binding Site Id: 2 Ala574,Trp577,Leu603,Met604,Pro541,Ala573, Gln570,Lys667, Arg569,Glu540,Ile539,Leu544, Ala607,Glu542,Tyr663,Arg611, Val543,Phe623, Trp610,Tyr660,Leu664,Glu537,Arg614,Lys576, Ala580,Val538,Ile572,Ala624,Leu566,Tyr545 |
Leu563 ,Asn564,Leu566, Gly567,Gln570, Trp600,Met601,Met604,Leu608, Arg611, Phe623,Leu732, Tyr735,Cys736, Thr739,Phe749,Leu753 |
Binding Site Id: 1 Leu563,Asn564,Gly567, Gln570,Met604,Arg611,Tyr735,Cys736,Thr739,Phe749
Binding Site Id: 2 Gln570,Met604,Arg611 |
|
|
ANDROGEN (PDB ID:3L3X) |
Leu704,Asn705,Gln711,Met745,Met749,Arg752,Thr877 |
Binding Site Id: 1 Asn705,Trp741,Met742,Thr877,Phe891,Met895, Ile899,Leu704, Pro892,Met780,Gln783,Cys784, Met787,Leu873,Phe764,Gln711, Met745,Val746, Met749,Gly708,Leu707,Phe876,Leu701,Leu880, Phe770 Binding Site Id: 2 Val684,Val685,Arg752,Tyr763,Ala765,Pro766, Gly683,Phe764, Val715,Leu744,Met745,Ala748, Lys808,Pro682,Gln711,His714 , Met749,Asn756, Trp718,Glu678,Glu681,Leu805,Trp751,Phe804, Pro801,Leu677,Cys686,Leu707,Thr755 |
Glu793,Trp796,Leu797, Arg854,Arg855, Gln858 , Lys861,Leu862,Ser865 |
Binding Site Id: 1 No Match
Binding Site Id: 2 No Match |
|
|
CARBOXY-IBUPROFEN (CID:10444113)
|
LDH-A (PDB ID:4ZVV) |
Gly28,Ala29,Val30,Asp51, Val52,Ile53, Thr94,Ala95,Gly96,Arg98,Ile115, Val135,Asn137,Trp147,Pro153,Lys154,Leu164, Asp165,Arg168,Arg170,His185,His192,Gly193, Asp194,Ala237,Tyr238,Ile241, Gly245,Thr247,Ile251 |
Binding Site Id: 1 Ser160,Gly161,Leu164,His192,Ile251,Val135,Val139, Asp165, Val25,Gly26,Val50,Asp51,Val52,Tyr82,Ile119, Lys80,Val30, Asn137,Ser136,Ala95,Phe118,Ile115, Thr247,Thr94,Arg168,Val27 ,Gly28,Ala29,Gly31, Ser248,Gly96,Gly193,Val233, Ser196,Pro138 ,Asp140, Glu191,Leu322 ,Ala237,Val234, Asp194,Ile141,Asn114, Ile53,Lys56 ,Tyr246,Ser236, Asn112,Ilea241,Ala97,Ser195, Thr321 ,Arg98,Val240 ,Tyr238 , Ile325,Gly245 ,Leu108, Val109,Arg105, Gln99,Glu239, Ser104,Asn107,Lys242,Glu103, Gln100,Ser318, Leu106,Glu328,Gly324,Asp320 Binding Site Id: 2 Trp249,Ala250,Leu253,Ala167,Leu164,Arg168,Ser236, Ala237, Val240,Tyr171,Leu172,Lys41,Asp257,Glu260,Lys244, Thr247, Ser248,Arg170,Pro181,Leu182,Asn163,Val269 , His270 ,Pro271, Trp187 ,Ser254,Ser166,Gly186,His185 , His180,Arg268,Asp165, Ile251,Val233,Gln232,Glu239,Glu235 |
Leu164,Ala167,Arg168, Tyr171,Val240, Lys244, Thr247,Ser248,Trp249, Ala250,Ile251 |
Binding Site ID: 1 Leu164,Arg168,Thr247,Ile251
Binding Site ID: 2 Leu164,Arg168,Thr247,Ile251
|
|
SURVIVIN (PDB ID:3UEC) |
Phe13,Leu14,Lys15,Asp16,Arg18,Gly30, Glu40,Glu51,Leu54,Cys57,Cys60,Lys62 ,Glu63, Leu64,Glu65,Gly66,Trp67,Asp70,Asp71, Glu76,His77,His80,Cys84,Phe86,Val89,Gln92, Glu94,Glu95,Thr97,Gly99,Glu107,Lys120,Glu123, Ala128,Val131,Arg132,Ala139,Asp142 |
Binding Site Id: 1 Phe13,Phe86,Val89,Phe93,Leu96,Leu104,Phe59,Lys91, Arg18,Leu14,Gln92,Glu94,Lys90,Lys15,Leu87,Ser88, Glu40,Ala85,Ala41,Glu95,Lys78,Asp16,Ile74,His17,Ile19 Binding Site Id: 2 Lys62,Lys115,Asn118,Lys122,Cys60,Ala114,Asn111, Ser81,Gln56,Phe61,Glu63,Asn119,His80, Leu64 ,Leu54,Glu51,Glu65
|
Phe13,Leu14,Lys15, Arg18, Glu40,Ala41,Ile74, Phe86,Leu87,Val89,Lys90 Lys91,Gln92,Phe93,Leu96 |
Binding Site Id: 1 Phe13,Leu14,Lys15,Arg18,Glu40,Phe86,Val89,Gln92
Binding Site Id: 2 No Match
|
|
|
GLUCOCORTICOIDS (PDB ID:4UDD) |
Glu540,Pro541,Met560 Leu563,Asn564,Gly567 ,Gln570,Trp577,Met604,Trp610, Arg611,Tyr640,Asp641,Gln642 His645,Met646,Tyr648 Tyr660, Lys667,Met691, Lys695, Val702,Ser708,Trp712,Phe715, Glu727,Asn731,Tyr735, Cys736,Gln738,Thr739,Met745, Ile747,Phe749, Gln776,Lys777 |
Binding Site Id: 1 Asn564,Trp600,Met601,Cys736,Phe749,Leu753, Leu563,Gly567,Thr739,Leu732,Tyr735,Met604, Gln570,Phe623,Met560,Ala605, Met646,Ile747, Gln642,Leu608,Leu566,Arg611, Cys643,Met639,Ile559,Ile629,Asp641,Asn731, Lys644,His645,Asn734,Gln738,Glu730,Met745, Thr556,Leu733,Thr562,Met565,Leu603,Ala607,Trp557 Binding Site Id: 2 Ala574,Trp577,Leu603,Met604,Pro541,Ala573, Gln570,Lys667, Arg569,Glu540,Ile539,Leu544, Ala607,Glu542,Tyr663,Arg611, Val543,Phe623, Trp610,Tyr660,Leu664,Glu537,Arg614,Lys576, Ala580,Val538,Ile572,Ala624,Leu566,Tyr545 |
Met560,Leu563,Asn564,Leu566,Gly567, Gln570,Trp600,Met601,Met604,Ala605, Leu608,Arg611,Phe623,Met646,Tyr735,Cys736, Phe749 |
Binding Site Id: 1 Met560,Leu563,Asn564,Gly567,Gln570,Met604,
Binding Site Id: 2 Gln570,Met604,Arg611 |
|
|
ANDROGEN (PDB ID:3L3X) |
Leu704,Asn705,Gln711,Met745,Met749,Arg752,Thr877 |
Binding Site Id: 1 Asn705,Trp741,Met742,Thr877,Phe891, Met895,Ile899,Leu704, Pro892,Met780, Gln783,Cys784,Met787,Leu873,Phe764,Gln711, Met745,Val746,Met749,Gly708,Leu707, Phe876,Leu701,Leu880, Phe770 Binding Site Id: 2 Val684,Val685,Arg752,Tyr763,Ala765,Pro766, Gly683,Phe764, Val715,Leu744,Met745,Ala748, Lys808,Pro682,Gln711,His714 , Met749,Asn756, Trp718,Glu678,Glu681,Leu805,Trp751, Phe804, Pro801,Leu677,Cys686,Leu707,Thr755 |
Glu793,Trp796,Leu797, Arg854, Arg855,Gln858, Lys861,Leu862,Ser865 |
Binding Site Id: 1 No Match
Binding Site Id: 2 No Match
|
|
|
2-HYDROXY-IBUPROFEN (CID:10443535) |
LDH-A (PDB ID:4ZVV) |
Gly28,Ala29,Val30,Asp51,Val52,Ile53,Thr94, Ala95,Gly96,Arg98,Ile115,Val135,Asn137,Trp147, Pro153,Lys154,Leu164,Asp165,Arg168,Arg170,His185, His192,Gly193,Asp194,Ala237,Tyr238, Ile241,Gly245,Thr247,Ile251 |
Binding Site Id: 1 Ser160,Gly161,Leu164,His192,Ile251,Val135,Val139, Asp165, Val25,Gly26,Val50,Asp51,Val52,Tyr82, Ile119,Lys80,Val30, Asn137,Ser136,Ala95, Phe118,Ile115,Thr247,Thr94,Arg168,Val27, Gly28,Ala29,Gly31,Ser248,Gly96,Gly193, Val233,Ser196,Pro138 ,Asp140,Glu191,Leu322 , Ala237,Val234,Asp194,Ile141,Asn114, Ile53,Lys56 , Tyr246,Ser236,Asn112,Ilea241,Ala97,Ser195,Thr321, Arg98,Val240,Tyr238,Ile325,Gly245,Leu108,Val109, Arg105, Gln99,Glu239,Ser104,Asn107,Lys242, Glu103,Gln100,Ser318, Leu106,Glu328,Gly324,Asp320 Binding Site Id: 2 Trp249,Ala250,Leu253,Ala167,Leu164,Arg168, Ser236,Ala237, Val240,Tyr171,Leu172,Lys41, Asp257,Glu260,Lys244,Thr247, Ser248,Arg170, Pro181,Leu182,Asn163,Val269 ,His270 ,Pro271, Trp187 ,Ser254,Ser166,Gly186,His185 ,His180, Arg268,Asp165, Ile251,Val233,Gln232,Glu239,Glu235 |
Arg168,Tyr171,Leu172, Ser236,Val240, Lys244, Thr247,Ser248,Trp249, Ala250,Ile251 |
Binding Site Id: 1 Arg168,Thr247,Ile251
Binding Site Id: 2 Arg168,Thr247,Ile251
|
|
SURVIVIN (PDB ID:3UEC) |
Phe13,Leu14,Lys15,Asp16,Arg18,Gly30,Glu40, Glu51,Leu54,Cys57,Cys60,Lys62,Glu63,Leu64, Glu65,Gly66,Trp67,Asp70,Asp71,Glu76,His77,His80, Cys84,Phe86,Val89,Gln92,Glu94,Glu95,Thr97,Gly99, Glu107,Lys120,Glu123,Ala128,Val131,Arg132,Ala139,Asp142 |
Binding Site Id: 1 Phe13,Phe86,Val89,Phe93,Leu96,Leu104,Phe59, Lys91,Arg18,Leu14,Gln92,Glu94,Lys90,Lys15,Leu87, Ser88,Glu40,Ala85,Ala41,Glu95, Lys78,Asp16,Ile74,His17,Ile19 Binding Site Id: 2 Lys62,Lys115,Asn118,Lys122,Cys60,Ala114, Asn111,Ser81,Gln56,Phe61,Glu63,Asn119, His80,Leu64 ,Leu54,Glu51,Glu65 |
Leu6 ,Pro7,Trp10,Phe93 ,Glu94,Leu96,Thr97, Leu98 ,Phe101 |
Binding Site Id: 1 Glu94
Binding Site Id: 2 No Match
|
|
|
GLUCOCORTICOIDS (PDB ID:4UDD) |
Glu540,Pro541,Met560,Leu563,Asn564,Gly567,Gln570, Trp577,Met604,Trp610,Arg611,Tyr640,Asp641,Gln642, His645,Met646,Tyr648,Tyr660,Lys667,Met691,Lys695, Val702 ,Ser708,Trp712,Phe715,Glu727,Asn731,Tyr735, Cys736,Gln738,Thr739,Met745,Ile747,Phe749,Gln776,Lys777 |
Binding Site Id: 1 Asn564,Trp600,Met601,Cys736,Phe749,Leu753, Leu563,Gly567,Thr739,Leu732,Tyr735, Met604,Gln570,Phe623,Met560,Ala605, Met646,Ile747,Gln642,Leu608,Leu566, Arg611,Cys643,Met639,Ile559,Ile629,Asp641, Asn731,Lys644,His645,Asn734,Gln738,Glu730, Met745,Thr556,Leu733,Thr562,Met565, Leu603,Ala607,Trp557 Binding Site Id: 2 Ala574,Trp577,Leu603,Met604,Pro541, Ala573,Gln570,Lys667, Arg569,Glu540, Ile539,Leu544,Ala607,Glu542,Tyr663,Arg611, Val543,Phe623,Trp610,Tyr660,Leu664,Glu537, Arg614,Lys576, Ala580,Val538,Ile572, Ala624,Leu566,Tyr545 |
Met560,Leu563,Asn564, Leu566,Gly567, Gln570,Trp600,Met604,Leu608,Arg611, Phe623,Tyr735,Thr739,Ile747,Phe749,Leu753 |
Binding Site Id: 1 Met560,Leu563,Asn564,Gly567,Gln570,Met604, Binding Site Id: 2 Gln570,Met604,Arg611
|
|
|
ANDROGEN (PDB ID:3L3X) |
Leu704,Asn705,Gln711,Met745,Met749,Arg752,Thr877 |
Binding Site Id: 1 Asn705,Trp741,Met742,Thr877,Phe891, Met895,Ile899,Leu704, Pro892,Met780, Gln783,Cys784,Met787,Leu873,Phe764, Gln711, Met745,Val746,Met749,Gly708, Leu707,Phe876,Leu701,Leu880, Phe770 Binding Site Id: 2 Val684,Val685,Arg752,Tyr763,Ala765, Pro766,Gly683,Phe764, Val715,Leu744, Met745,Ala748,Lys808,Pro682,Gln711, His714 , Met749,Asn756,Trp718, Glu678,Glu681,Leu805,Trp751, Phe804, Pro801,Leu677,Cys686,Leu707,Thr755 |
Glu793,Trp796,Leu797, Gln858,Lys861,Leu862, Ser865 |
Binding Site Id: 1 No Match
Binding Site Id: 2 No Match
|
|
|
IBUPROFEN METHYL ESTER (CID: 109101) |
LDH-A (PDB ID:4ZVV) |
Gly28,Ala29,Val30,Asp51, Val52,Ile53,Thr94,Ala95, Gly96,Arg98,Ile115,Val135, Asn137,Trp147,Pro153, Lys154,Leu164,Asp165,Arg168, Arg170,His185,His192, Gly193,Asp194,Ala237,Tyr238, Ile241,Gly245,Thr247,Ile251 |
Binding Site Id: 1 Ser160,Gly161,Leu164,His192,Ile251, Val135,Val139,Asp165, Val25,Gly26, Val50,Asp51,Val52,Tyr82,Ile119,Lys80, Val30, Asn137,Ser136,Ala95,Phe118, Ile115,Thr247,Thr94,Arg168,Val27, Gly28,Ala29,Gly31,Ser248,Gly96, Gly193,Val233,Ser196,Pro138 ,Asp140, Glu191,Leu322 ,Ala237,Val234,Asp194, Ile141,Asn114, Ile53,Lys56 ,Tyr246, Ser236,Asn112,Ilea241,Ala97,Ser195, Thr321 ,Arg98,Val240 ,Tyr238 ,Ile325, Gly245 ,Leu108,Val109,Arg105, Gln99, Glu239,Ser104,Asn107,Lys242,Glu103, Gln100,Ser318, Leu106,Glu328,Gly324,Asp320 Binding Site Id: 2 Trp249,Ala250,Leu253,Ala167,Leu164, Arg168,Ser236,Ala237, Val240,Tyr171, Leu172,Lys41,Asp257,Glu260,Lys244, Thr247, Ser248,Arg170,Pro181,Leu182, Asn163,Val269 ,His270 ,Pro271, Trp187 , Ser254,Ser166,Gly186,His185 ,His180, Arg268,Asp165, Ile251,Val233,Gln232,Glu239,Glu235 |
Val30,Thr94,Ala95,Gly96,Ala97,Leu108, Asn112, Val135,Ser136,Asn137, Leu164,His192,Thr247, Ile251 |
Binding Site Id: 1 Val30,Thr94,Ala95,Gly96,Val135, Asn137,Leu164,His192,Thr247,Ile251
Binding Site Id: 2 Leu164,Thr247,Ile251
|
|
SURVIVIN (PDB ID:3UEC) |
Phe13,Leu14,Lys15,Asp16, Arg18,Gly30,Glu40,Glu51, Leu54,Cys57,Cys60,Lys62,Glu63, Leu64,Glu65,Gly66, Trp67,Asp70,Asp71,Glu76,His77,His80, Cys84,Phe86,Val89, Gln92,Glu94,Glu95,Thr97,Gly99,Glu107, Lys120,Glu123Ala128, Val131,Arg132,Ala139,Asp142 |
Binding Site Id: 1 Phe13,Phe86,Val89,Phe93,Leu96,Leu104, Phe59,Lys91,Arg18,Leu14,Gln92,Glu94, Lys90,Lys15,Leu87,Ser88,Glu40, Ala85,Ala41,Glu95,Lys78,Asp16,Ile74,His17,Ile19 Binding Site Id: 2 Lys62,Lys115,Asn118,Lys122,Cys60, Ala114,Asn111,Ser81,Gln56,Phe61, Glu63,Asn119,His80,Leu64 ,Leu54,Glu51,Glu65 |
Phe59 ,Ala85 ,Ser88, Val89,Lys90, Lys91,Leu96,Glu100, Lys103,Leu104, Glu107 |
Binding Site Id: 1 Val89
Binding Site Id: 2 No Match
|
|
|
GLUCOCORTICOIDS (PDB ID:4UDD) |
Glu540,Pro541,Met560,Leu563,Asn564, Gly567,Gln570,Trp577,Met604,Trp610,Arg611, Tyr640,Asp641,Gln642,His645, Met646,Tyr648,Tyr660,Lys667,Met691,Lys695, Val702,Ser708,Trp712,Phe715,Glu727,Asn731, Tyr735,Cys736,Gln738,Thr739, Met745,Ile747,Phe749,Gln776,Lys777 |
Binding Site Id: 1 Asn564,Trp600,Met601,Cys736,Phe749, Leu753,Leu563,Gly567,Thr739,Leu732, Tyr735,Met604,Gln570,Phe623,Met560, Ala605, Met646,Ile747,Gln642,Leu608,Leu566, Arg611,Cys643,Met639,Ile559,Ile629, Asp641,Asn731,Lys644,His645,Asn734, Gln738,Glu730,Met745,Thr556,Leu733, Thr562,Met565,Leu603,Ala607,Trp557 Binding Site Id: 2 Ala574,Trp577,Leu603,Met604, Pro541,Ala573,Gln570,Lys667, Arg569,Glu540,Ile539,Leu544, Ala607,Glu542,Tyr663,Arg611, Val543,Phe623,Trp610,Tyr660, Leu664,Glu537,Arg614,Lys576, Ala580,Val538,Ile572,Ala624, Leu566,Tyr545 |
Thr556,Ile559,Met560,Leu563, Asn564,Met601, Phe623,Ile629,Met639,Gln642, Cys643,Met646, Leu732,Tyr735,Cys736,Thr739,Phe749 |
Binding Site Id: 1 Met560,Leu563,Asn564,Gln642, Met646,Tyr735,Cys736,Thr739,Phe749
Binding Site Id: 2 No Match |
|
|
ANDROGEN (PDB ID:3L3X) |
Leu704,Asn705,Gln711 Met745,Met749,Arg752 Thr877 |
Binding Site Id: 1 Asn705,Trp741,Met742,Thr877, Phe891,Met895,Ile899,Leu704, Pro892,Met780,Gln783,Cys784, Met787,Leu873,Phe764,Gln711, Met745,Val746,Met749,Gly708, Leu707,Phe876,Leu701,Leu880, Phe770 Binding Site Id: 2 Val684,Val685,Arg752,Tyr763, Ala765,Pro766,Gly683,Phe764, Val715,Leu744,Met745,Ala748, Lys808,Pro682,Gln711,His714 , Met749,Asn756,Trp718,Glu678, Glu681,Leu805,Trp751,Phe804, Pro801,Leu677,Cys686,Leu707,Thr755 |
Glu793,Trp796,Leu797, Arg854,Gln858,Lys861, Leu862,Ser865 |
Binding Site Id: 1 No Match
Binding Site Id: 2 No Match |
4.a
4.b
4.c
4.d
4.e
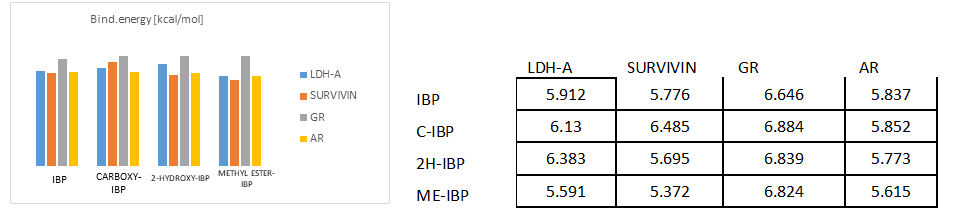
Fig. 4.a)-4.e) Drug-receptor docking analysis results with PatchDock and YASARA
Table 2. Results obtained from RAMPAGE statistics, ResProx, ERRAT, and PDBSum Server
|
Parameters |
LDH-A |
Survivin |
GR |
AR |
|
Resolution ((Å) |
2.2 |
2.18 |
1.8 |
1.55 |
|
RAMPAGE-Number of residues in favoured region (~98.0% expected) |
98.2%
|
98.5%
|
98.8%
|
98.8%
|
|
RAMPAGE-Number of residues in allowed region ( ~2.0% expected) |
1.8%
|
98.5%
|
1.2%
|
1.2%
|
|
RAMPAGE-Number of residues in outlier region |
0%
|
0%
|
0.0%
|
0.0%
|
|
ResProx-GOOD(0-1.5),MIDDLE(1.5-2.5), BAD (>2.5) |
2.159 |
1.918 |
1.526 |
2.075 |
|
ERRAT AT >91 |
97.4684 |
98.4615 |
100 |
96.281 |
|
PDBSum-Most favoured regions |
92.8% |
92.6% |
94.3% |
95.2% |
|
PDBSum-Additional allowed regions |
7.2% |
7.4% |
5.7% |
4.8% |
|
PDBSum-Generously allowed regions |
0% |
0% |
0.0% |
0.0% |
|
PDBSum-Disallowed regions |
0% |
0% |
0.0% |
0.0% |
Table 3. Results obtained from VADAR analysis
|
|
Mean H-bond distance |
Mean H-bond energy |
Residue with H-bonds |
Helix
|
Beta
|
Coil
|
Turn
|
|||
|
Protein Name |
Observed |
Expected |
Observed |
Expected |
Observed |
Expected |
|
|
|
|
| LDH-A | 2.2 sd=0.3 | 2.2 sd=0.4
|
-1.7 sd=0.9 | -2.0 sd=0.8
|
262 (79%) | 248 (75%)
|
142 (42%) | 67 (20%) | 122(36%)
|
88 (26%)
|
| Survivin | 2.2 sd=0.4 | 2.2 sd=0.4
|
-1.8 sd=1.0 | -2.0 sd=0.8
|
112 (81%) | 103 (75%)
|
69 (50%)
|
16 (11%)
|
53 (38%)
|
40 (28%)
|
| GR | 2.2 sd=0.3 | 2.2 sd=0.4
|
-1.7 sd=1.0 | -2.0 sd=0.8
|
211 (84%) | 186 (75%)
|
170 (68%)
|
14 (5%)
|
65 (26%)
|
44 (17%)
|
| AR | 2.2 sd=0.4 | 2.2 sd=0.4
|
-1.7 sd=1.0 | -2.0 sd=0.8
|
218 (87%) | 187 (75%)
|
170 (68%)
|
14 (5%)
|
66 (26%)
|
32 (12%)
|
Table 4. Results obtained from Lipinski filter
|
Parameters |
IBP |
Carboxy-IBP |
Hydroxy-IBP |
IBP methyl ester |
|
Molecular mass less than 500 Dalton |
205 |
234 |
221 |
220 |
|
High lipophilicity (expressed as LogP less than 5) |
1.7385 |
-0.5315 |
0.8534 |
3.1615 |
|
Hydrogen bond donors(Lessthan5) |
0 |
0 |
1 |
0 |
|
Hydrogen bond acceptors( Lessthan10) |
2 |
4 |
3 |
2 |
|
Molar refractivity should be between 40-130 |
58.405987
|
57.738991 |
59.865788 |
65.414986 |
Table 5. Results obtained from admetSAR
|
Parameters |
IBP |
Carboxy-IBP |
Hydroxy-IBP |
IBP methyl ester |
|
Blood-Brain Barrier |
BBB+ |
BBB+ |
BBB+ |
BBB+ |
|
Human Intestinal Absorption |
HIA+ |
HIA+ |
HIA+ |
HIA+ |
|
Caco-2 Permeability |
Caco2+ |
Caco2+ |
Caco2+ |
Caco2+ |
|
P-glycoprotein Substrate |
Non-substrate |
Non-substrate |
Non-substrate |
Non-substrate |
|
AMES Toxicity |
Non AMES toxic |
Non AMES toxic |
Non AMES toxic |
Non AMES toxic |
|
Carcinogens |
Carcinogens |
Non-carcinogens |
Non-carcinogens |
Carcinogens |
|
Acute Oral Toxicity |
III |
III |
III |
III |
|
Rat Acute Toxicity |
2.3092LD50,mol/kg |
2.0465 LD50,mol/kg |
1.8844LD50,mol/kg |
1.9786LD50,mol/kg |
|
CYP450 Substrate & Inhibitor |
Non-substrate, Non-inhibitor |
Non-substrate, Non-inhibitor |
Non-substrate, Non-inhibitor |
Non-substrate, Non-inhibitor |
|
hERG |
Weak inhibitor |
Weak inhibitor |
Weak inhibitor |
Weak inhibitor |
Table 6. Results obtained from FAFDrugs4
|
Parameters |
IBP |
Carboxy-IBP |
Hydroxy-IBP |
IBP methyl ester |
|
Veber Rule |
Good |
Good |
Good |
Good |
|
Egan Rule |
Good |
Good |
Good |
Good |
|
Solubility (mg/l) |
6613.10 |
18409.52 |
15552.37 |
7082.91 |
|
n_LipinskiViolations |
0 |
0 |
0 |
0 |
|
Rotatable Bonds |
4 |
5 |
4 |
5 |
|
Rigid Bonds |
7 |
8 |
7 |
7 |
|
n_carbon |
13 |
13 |
13 |
14 |
|
ratioH/C |
0.15 |
0.31 |
0.23 |
0.14 |
|
NumCharges |
1 |
2 |
1 |
0 |
|
logP |
3.89 |
2.29 |
2.50 |
3.87 |
|
tPSA |
40.13 |
80.26 |
60.36 |
26.3 |
|
State |
Accepted |
Accepted |
Accepted |
Accepted |
Table 7. Results obtained from SwissADME
|
Parameters |
IBP |
Carboxy-IBP |
Hydroxy-IBP |
IBP methyl ester |
|
SMILES |
CC(Cc1ccc(cc1)[C@H](C(=O)O)C)C |
OC(=O)C(Cc1ccc(cc1)C(C(=O)O)C)C |
CC(c1ccc(cc1)CC(O)(C)C)C(=O)O |
COC(=O)C(c1ccc(cc1)CC(C)C)C |
|
Formula |
C13H18O2 |
C13H16O4 |
C13H18O3 |
C14H20O2 |
|
Molecular weight |
206.28 g/mol |
236.26 g/mol |
222.28 g/mol |
220.31 g/mol |
|
Num. heavy atoms |
15 |
17 |
16 |
16 |
|
Consensus Log Po/w |
3.01 |
2.01 |
2.11 |
3.44 |
|
Lipinski |
Yes; 0 violation |
Yes; 0 violation |
Yes; 0 violation |
Yes; 0 violation |
|
Bioavailability Score |
0.56 |
0.56 |
0.56 |
0.55 |
|
PAINS |
0 alert |
0 alert |
0 alert |
0 alert |
|
Brenk |
0 alert |
0 alert |
0 alert |
0 alert |
|
Leadlikeness |
No; 1 violation: MW<250 |
No; 1 violation: MW<250 |
No; 1 violation: MW<250 |
No; 2 violations: MW<250, XLOGP3>3.5 |
|
Synthetic accessibility |
1.92 |
2.31 |
1.95 |
2.16 |
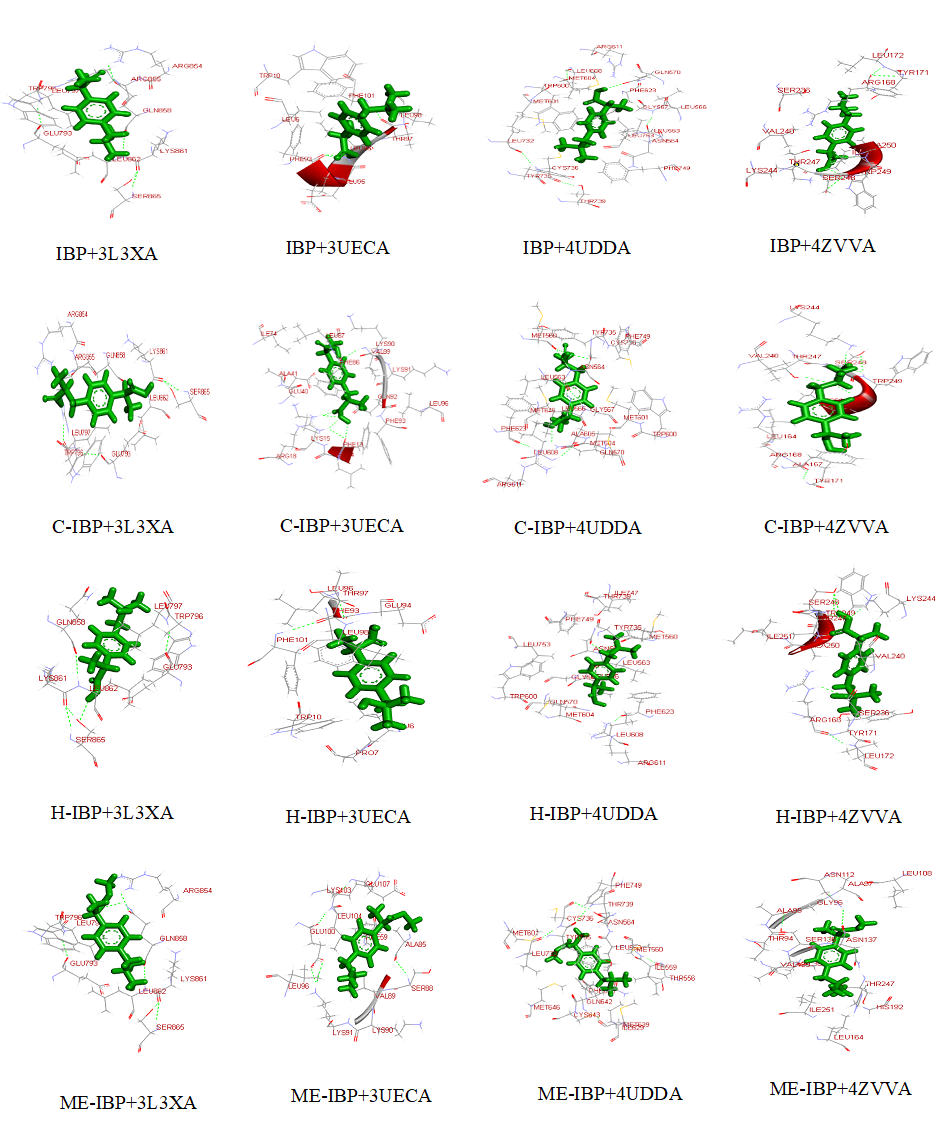
Fig 5. Visualization of docked complexes. The figure shows 3-D models of docked complexes as visualized by Discovery Studio 3.5, showing interactions of IBP and its derivatives with the target receptors.
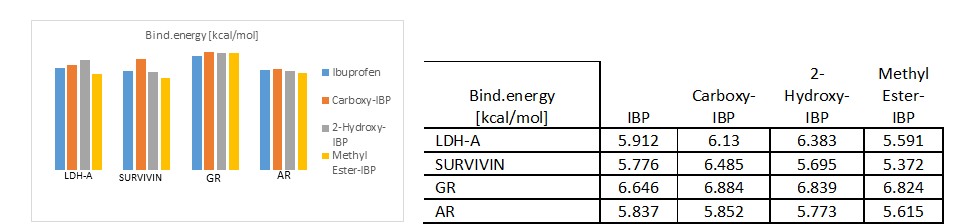
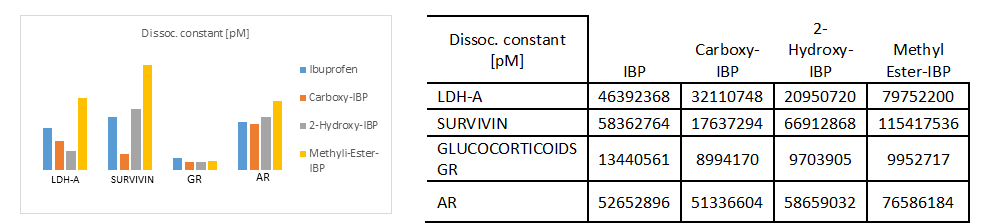
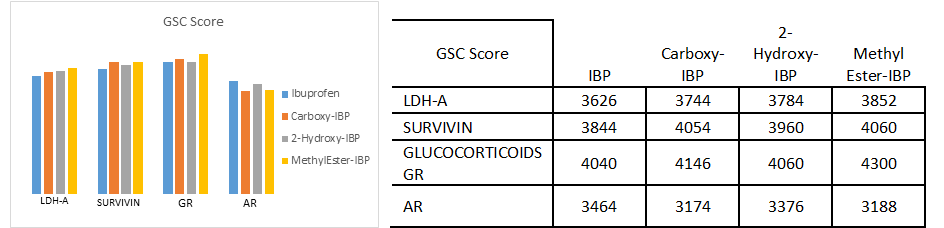
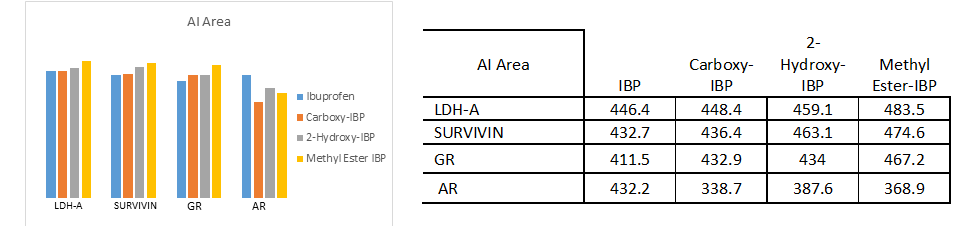
The docking analysis result to PatchDock server and YASARA software are shown in Fig. 4.a)-e) which are reported to be highly reliable tools for molecular docking studies (Krieger et al., 2014). The docking analysis was based on the vital parameters of molecular docking indicating the strength of molecular interactions (Yadav et al., 2017). Fig 4.a) and 4.b) shows a graphical representation of the Binding energy (in kcal/mol) and Dissociation constant (in pM) respectively between IBP and its derivatives and the receptor molecules using YASARA software. Fig 4.c) and 4.d) is a graphical representation of GSC Score and AI Area respectively between IBP and its derivatives and the receptor molecules using PatchDock server. The docked complexes are visualized by Discovery Studio 3.5 and are shown in Fig 5. The optimum docking and better binding is indicated by the lower value of Dissociation constant and more positive values of Binding energy and higher values of GSC Score and AI Area (Rocheleau et al., 2016; Pandey et al., 2019).From the observed values Glucocorticoids Receptor (GR) shows the minimum value in dissociation constant (Kd) and higher value of binding energy, GSC Score and AI Area with all the ligands when all the receptor-ligand interaction is compared Fig 4.e). And among the drugs the Ibuprofen derivatives- Carboxy IBP and 2-Hydroxy IBP shows the best result with minimum value in dissociation constant (Kd) and higher value of binding energy, GSC Score and AI Area.
The Table 2. represents the results from RAMPAGE statistics, ResProx, ERRAT and PDBSum server showing all the receptor within the acceptable range. Table 3.shows the VADAR analysis with the hydrogen bond statistics. Table 4.shows zero Lipinski violation for all the ligands and Table 6.and Table 7.shows the FAFDrugs4 and SwissADME results respectively. Table 5.shows the admetSAR properties where Carboxy-IBP and 2-Hydroxy-IBP are non-carcinogenic and rest all the parameters are within the range.
Based on all the docking calculations and druglikeness analysis finally it can be concluded that Ibuprofen derivatives- Carboxy IBP and 2-Hydroxy IBP shows the best result with minimum value in dissociation constant (Kd) and higher value of binding energy, GSC Score and AI Area and among all the targets Glucocorticoids Receptor (GR) shows the best result with lowest dissociation constant and highest binding energy, GSC Score and AI Score which can be summarized and understood from the graphical representations in Fig. 4.a)-d).GR shows the best docking interaction with Ibuprofen derivatives Carboxy-Ibuprofen and 2-Hydroxy-Ibuprofen, which are non-carcinogenic, better and more efficacious than the parent drug Ibuprofen itself. Glucocorticoids Receptor (GR) can be considered a novel receptor for the Ibuprofen derivatives. Ibuprofen derivatives Carboxy IBP and 2-Hydroxy IBP are more efficacious from the observations can be used for cancer therapeutics like breast, lung and can be utilized for further research.
I would like to thank Dr. Vinay Kumar Singh, Information officer of Centre for Bioinformatics, School of Biotechnology, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India PIN-221005 for his continuous support, guidance and sharing his pearl of wisdom with me during the course of this research.
Ambrosini, G., Adida, C., & Altieri, D. C. (1997). A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nature medicine, 3(8), 917-921. PMid:9256286
View Article PubMed/NCBIBakker, E., Tian, K., Mutti, L., Demonacos, C., Schwartz, J. M., & Krstic-Demonacos, M. (2017). Insight into glucocorticoid receptor signalling through interactome model analysis. PLoS computational biology, 13(11), e1005825. PMid:29107989
View Article PubMed/NCBIBittoni,M.A.,Carbone,D.P.,&Harris,R.E.(2017).Ibuprofenandfatallungcancer:Abrief report of the prospective results from the Third National Health and Nutrition Examination Survey (NHANES III). Molecular and clinical oncology, 6(6), 917-920. PMid:28588790
View Article PubMed/NCBIConzen, S. D. (2017). Recent Advances in Understanding Glucocorticoid Receptor Function in Cancer. Clinical advances in hematology & oncology: H&O, 15(5), 338.
Di Stefano, G., Manerba, M., Di Ianni, L., & Fiume, L. (2016). Lactate dehydrogenase inhibition: exploring possible applications beyond cancer treatment. Future medicinal chemistry, 8(6), 713-725. PMid:27054686
View Article PubMed/NCBIDohi,T.,Beltrami,E., Wall,N.R.,Plescia,J.,&Altieri,D.C.(2004).Mitochondrialsurvivin inhibits apoptosis and promotes tumorigenesis. The Journal of clinical investigation, 114(8), 1117-1127. PMid:15489959
View Article PubMed/NCBIFriedrichs,B.,Siegel,S.,HaldAndersen,M.,Schmitz,N.,&Zeis,M.(2006).Survivin-derived peptide epitopes and their role for induction of antitumor immunity in hematological malignancies. Leukemia & lymphoma, 47(6), 978-985. PMid:16840186
View Article PubMed/NCBIFujita, K., & Nonomura, N. (2019). Role of androgen receptor in prostate cancer: a review. The world journal of men's health, 37(3), 288-295. PMid:30209899
View Article PubMed/NCBIGiardiello, F. M., Offerhaus, G. J. A., & DuBois, R. N. (1995). The role of nonsteroidal anti- inflammatory drugs in colorectal cancer prevention. European Journal of Cancer, 31(7-8), 1071-1076. 00137-8
View ArticleGreenspan, E. J., Madigan, J. P., Boardman, L. A., & Rosenberg, D. W. (2011). Ibuprofen inhibits activation of nuclear β-catenin in human colon adenomas and induces the phosphorylation of GSK-3β. Cancer Prevention Research, 4(1), 161-171. PMid:21205744
View Article PubMed/NCBIGurpinar,E.,Grizzle,W.E.,&Piazza,G.A.(2013).COX-independentmechanismsofcancer chemoprevention by anti-inflammatory drugs.Frontiers in oncology,3,181. PMid:23875171
View Article PubMed/NCBIHaché, R. J., Tse, R., Reich, T., Savory, J. G., & Lefebvre, Y. A. (1999). Nucleocytoplasmic trafficking of steroid-free glucocorticoid receptor. Journal of Biological Chemistry, 274(3), 1432-1439. PMid:9880517
View Article PubMed/NCBIHarris, R. E., Beebe-Donk, J., Doss, H., & Doss, D. B. (2005). Aspirin, ibuprofen, and other non-steroidal anti-inflammatory drugs in cancer prevention: a critical review of non-selective COX-2 blockade. Oncology reports, 13(4), 559-583. PMid:15756426
View Article PubMed/NCBIHur, E., Pfaff, S. J., Payne, E. S., Grøn, H., Buehrer, B. M., & Fletterick, R. J. (2004). Recognition and accommodation at the androgen receptor coactivator binding interface. PLoS biology, 2(9). PMid:15328534
View Article PubMed/NCBIJanssen, A., Maier, T. J., Schiffmann, S., Coste, O., Seegel, M., Geisslinger, G., & Grösch, S. (2006). Evidence of COX-2 independent induction of apoptosis and cell cycle block inhuman colon carcinoma cells after S-or R-ibuprofen treatment. European journal of pharmacology, 540(1-3), 24-33. PMid:16730702
View Article PubMed/NCBIKach, J., Conzen, S. D., & Szmulewitz, R. Z. (2015). Targeting the glucocorticoid receptor in breast and prostate cancers. Science translational medicine, 7(305), 305ps19-305ps19. PMid:26378243
View Article PubMed/NCBIKrieger, E., & Vriend, G. (2014). YASARA View-molecular graphics for alldevices-from smartphones to workstations. Bioinformatics, 30(20), 2981-2982. PMid:24996895
View Article PubMed/NCBILeidgens, V., Seliger, C., Jachnik, B., Welz, T., Leukel, P., Vollmann-Zwerenz, A., & Hau,P. (2015). Ibuprofen and diclofenac restrict migration and proliferation of human glioma cells by distinct molecular mechanisms. PloS one, 10(10). PMid:26485029
View Article PubMed/NCBIMargulis,V.,Lotan,Y.,&Shariat,S.F.(2008).Survivin:apromisingbiomarkerfordetection and prognosis of bladder cancer.World journal of urology,26(1),59-65. PMid:17962949
View Article PubMed/NCBIMiyamoto, H., Altuwaijri, S., & Chang, C. (2008). Androgen receptor in prostate cancer progression. In Prostate Cancer (pp. 129-146). Humana Press.
View ArticleOuyang, N., Ji, P., & Williams, J. L. (2013). A novel NSAID derivative, phospho-ibuprofen, preventsAOM-inducedcoloncancerinrats.Internationaljournalofoncology,42(2),643-650. PMid:23291777
View Article PubMed/NCBIPandey, S. K., Yadav, S., Goel, Y., Temre, M. K., Singh, V. K., & Singh, S. M. (2019). Molecular docking of anti-inflammatory drug diclofenac with metabolic targets: Potential applications in cancer therapeutics. Journal of theoretical biology, 465, 117-125. PMid:30653975
View Article PubMed/NCBIRatman, D., Berghe, W. V., Dejager, L., Libert, C., Tavernier, J., Beck, I. M., & DeBosscher,K. (2013). How glucocorticoid receptors modulate the activity of other transcription factors: a scope beyond tethering. Molecular and cellular endocrinology, 380(1-2), 41-54. PMid:23267834
View Article PubMed/NCBIRocheleau, A. D., Cao, T. M., Takitani, T., & King, M. R. (2016). Comparison of human and mouse E-selectin binding to Sialyl-Lewis x. BMC structural biology, 16(1),10. PMid:27368167
View Article PubMed/NCBISharma, B. B., Kalia, P., Yadava, D. K., Singh, D., & Sharma, T. R. (2016). Genetics and molecular mapping of black rot resistance locus Xca1bc on chromosome B-7 in Ethiopian mustard (Brassica carinata A. Braun). PloS one, 11(3). PMid:27023128
View Article PubMed/NCBISheng, S. L., Liu, J. J., Dai, Y. H., Sun, X. G., Xiong, X. P., & Huang, G. (2012).Knockdown of lactate dehydrogenase A suppresses tumor growth and metastasis of human hepatocellular carcinoma. The FEBS journal, 279(20), 3898-3910. PMid:22897481
View Article PubMed/NCBIShi, M., Cui, J., Du, J., Wei, D., Jia, Z., Zhang, J., ... & Xie, K. (2014). A novel KLF4/LDHA
A Novel KLF4/LDHA Signaling Pathway Regulates Aerobic Glycolysis in and Progression of Pancreatic Cancer.Clinical Cancer Research, 20(16), 4370-4380. PMid:24947925
View Article PubMed/NCBISundahl, N., Clarisse, D., Bracke, M., Offner, F., Berghe, W. V., & Beck, I. M. (2016). Selective glucocorticoid receptor-activating adjuvant therapy in cancer treatments. Oncoscience, 3(7-8), 188.
Tamm,I.,Wang,Y.,Sausville,E.D.,Scudiero,D.A.,Vigna,N.,Oltersdorf,T.,&Reed,J.C. (1998). IAP-family protein survivin inhibits caspase activity and apoptosis induced by Fas (CD95), Bax, caspases, and anticancer drugs. Cancer research, 58(23),5315-5320.
Thun, M. J., Henley, S. J., & Patrono, C. (2002). Nonsteroidal anti-inflammatory drugs as anticancer agents: mechanistic, pharmacologic, and clinical issues. Journal of the National Cancer Institute, 94(4), 252-266. PMid:11854387
View Article PubMed/NCBIVandevyver, S., Dejager, L., & Libert, C. (2012). On the trail of the glucocorticoid receptor: into the nucleus and back. Traffic, 13(3), 364-374. PMid:21951602
View Article PubMed/NCBIXian, Z. Y., Liu, J. M., Chen, Q. K., Chen, H. Z., Ye, C. J., Xue, J., ... & Kuang, S. J. (2015).
Inhibition of LDHA suppresses tumor progression in prostate cancer. Tumor Biology, 36(10), 8093-8100. PMid:25983002 PMCid:PMC4605959
View Article PubMed/NCBIYadav,S.,Pandey,S.K.,Singh,V.K.,Goel,Y.,Kumar,A.,&Singh,S.M.(2017).Molecular docking studies of 3-bromopyruvate and its derivatives to metabolic regulatory enzymes: Implication in designing of novel anticancer therapeutic strategies. PloS one, 12(5). PMid:28463978
View Article PubMed/NCBIZhao, Y. H., Zhou, M., Liu, H., Ding, Y., Khong, H. T., Yu, D., ... & Tan, M. (2009).
Upregulation of lactate dehydrogenase A by ErbB2 through heat shock factor 1 promotes breast cancer cell glycolysis and growth. Oncogene, 28(42), 3689-3701. PMid:19668225
View Article PubMed/NCBI