Abbas Khan
Email: abbaskhan@sjtu.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 1
Page No: 328-342
Abbas Khan
Email: abbaskhan@sjtu.edu.cn
Shoaib Saleem1, Abbas Khan2*, Syed Shujait Ali2, Abrar Mohammad Sayaf2, Mazhar Khan3,
1National Center for Bioinformatics, Quaid-e-Azam University, Islamabad, Pakistan
2Center for Biotechnology and Microbiology, University of Swat, Pakistan
3The CAS Key Laboratory of Innate Immunity and Chronic Diseases, Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, CAS Center for Excellence in Molecular Cell Science, University of Science and Technology of China (USTC), Collaborative Innovation Center of Genetics and Development, Hefei, 230027, Anhui, China
Shoaib Saleem: shoaibsaleem90@gmail.com; Syed Shujait Ali: shujaitswati@uswat.edu.pk; Abrar Mohammad Sayaf: Chemdr07@gmail.com; Mazhar Khan: mazharsw@mail.ustc.edu.cn
Xiaoqiang Sun(sunxq6@mail.sysu.edu.cn)
Kaue Santana da Costa(kaue.costa@ufopa.edu.br)
Xin Chen(chenxin_0001@126.com)
Reza Ahangari Cohan(cohan_r@yahoo.com)
Shoaib Saleem, Abbas Khan, Syed Shujait Ali, Abrar Mohammad Sayaf, Mazhar Khan,Molecular modeling of the multiple-drug resistant protein (MRP7) and pharmacophore modelling based virtual screening to identify novel drugs against cancer(2020)Journal of Computational Chemistry & Molecular Modeling 4(1) p:328-342
Chemotherapy is extensively used for the treatment of various types of cancer. Multidrug resistance (MDR) against chemotherapeutics agents remains one of the most important hurdle in the successful chemotherapy of cancer. The efflux mechanism of ABC transporters is considered as the primary cause of Multidrug resistance (MDR). CC10 (MRP7) is recently described as one of the new players in the development of MDR in cancer cells. Therefore, we used a computational approach to model the 3D structure of ABCC10 and used the already reported anti-cancerous compounds against the ligand binding site of ABCC10. In this work, we have developed homology models of the ABCC10 transporter and assessed them in virtual screening for the identification of novel ligands. The models were generated by MOE, MODELLER, I-TASSER, EXPASY, PHYRE2. Energy minimization was carried out by using YASARA energy minimization server. The final model was built by combining all these models using MODELLER. The binding site was identified using MOE. The homology models were validated using different servers including Errat, Rampage, ProQ, and TM-Scoring. This approval was a confident way to dock known ligands against the binding pocket of ABCC10. The known ligands were docked, and Gemcitabine and Methotrexate were found to have good docking score. We used Gemcitabine as pharmacophoric input to identify novel hits from Cambridge database. A total of 5,151 hits were identified and enlisted. This study provides an insight into the knowledge of ABC transporter family inhibitors discovery.
Keywords: Molecular modeling,MRP7, pharmacophore modelling
Chemical therapy is widely utilized for the treatment of multiple types of cancer. Cell proliferation, growth and spread of malignant cells can be effectively controlled by using different chemotherapeutic agents. Resistance conferred to the chemical drugs is a substantial factor that confines the strength of chemotherapy and causes failure of cancer treatment (Michael M Gottesman, Fojo, & Bates, 2002). Overexpression of ABC transporters on the surface of membranes of cancer cells is correlate with Multidrug resistance (MDR). Multidrug resistance (MDR) against chemotherapeutics agents remains one of the most important hurdle in the successful chemotherapy of cancer. To date, 49 different ABCs are identified of which 14 are associated with different human diseases (Eckford & Sharom, 2008);(Kimura, Morita, Matsuo, & Ueda, 2007). The main differences among ABC transporters are observed in their substrate specificity, localization, and function and molecular mechanism of resistance (Higgins, 2007). Due to genome sequence similarities, ABC transporters have been assembled into seven subfamilies. The seven families include A to G (Dean & Annilo, 2005). P-gp which is also known as ABCB1, ABCC or MRP subfamily and ABCG2 or breast cancer resistance proteins (BCRP) are primarily observed the development of MDR in cancer cells (M. M. Gottesman & Ambudkar, 2001; Schinkel & Jonker, 2003).
ABC C family or MRP is one of the subfamilies among ATP binding cassette transporters which comprises of 9 members from Multiple Resistant Proteins 1 to Multiple Resistant Proteins 9. The MRPs represent 12 members of the C subfamily of ATP binding cassette transporters (Z. S. Chen & Tiwari, 2011). In 2001, ABCC10 was found a new member of ATP-binding cassette transporters family (Hopper et al., 2001). And its role in the expansion of MDR in cancer cells (G. D. Kruh, Guo, Hopper-Borge, Belinsky, & Chen, 2007). The human MRP7 gene ABCC10 is located on chromosome 6p21 (Hopper et al., 2001; Kruh et al., 2007). ABCC10 encode a protein of 171-kDa that consist of three MSD and two NBD (Hopper-Borge, Chen, Shchaveleva, Belinsky, & Kruh, 2004). Like other MRPs, ABCC10 is similarly related to C family ABC transporters involved in the directive of ion transport (Deeley, Westlake, & Cole, 2006; Gary D Kruh & Belinsky, 2003). The presence of ABCC10 was reported early as a factor of resistance to drugs. ABCC10, ABCC1, ABCC2, ABCC3, and ABCC6 are group of important ABC transporters located on the basolateral cell surface (Z.-S. Chen et al., 2003; E. Hopper-Borge, Chen, Shchaveleva, Belinsky, & Kruh, 2004; E. A. Hopper-Borge et al., 2011; Malofeeva, Domanitskaya, Gudima, & Hopper-Borge, 2012). The expression of ABCC10 has already been reported in ABCC10 in different organs of the body such as kidneys, heart, and brain. However low level of expression was also found in pancreas, ovaries, lymph nodes, liver, placenta, leukocytes, lungs, colon, and heart (Takayanagi et al., 2004).
Phylogenetically Multiple Resistant Proteins 7 is involved in the regulation of ion channels and also related to lipophilic anion pumps. It is concluded that ABCC10 is involved in phase III of detoxification and is one of the lipophilic anion transporters (Z.-S. Chen et al., 2003). In 2004 resistance to anticancer drugs posed by ABCC10 include vinblastine, vincristine, paclitaxel and docetaxel (E. Hopper-Borge et al., 2004) was reported. The highest level of expression of ABCC10 gene was found in pancreas(Takayanagi et al., 2004). It was also reported that derived peptide of ABCC10 can be used as immunoregulator and pose resistant to docetaxel (Naramoto et al., 2007). However resistant to vinorelbine in non-small cell lung cancer has already been reported (Wooden, Kalb, Cotter, & Soloski, 2005).
Resistance of epothilone B to ABCB1 and ABCC1, ABCC10 also been reported (Shen et al., 2009). Nilotinib, BCR-Abl tyrosine kinase inhibitors and Imatinib are reported as the potent inhibitors of the efflux function of ABCC10 efflux transporter (Shen et al., 2009). Sildenafil and Vardenafil, Phosphodiesterase 5 inhibitors reverse MDR mediated by ABCC10 (J. J. Chen et al., 2012). A distinctive feature has been marked to MRP7 to pose 9 to 13-fold resistance to a microtubule stabilizing agent docetaxel. It has also been reported that 3 to 4 fold level of resistance was detected to taxanes including vincristine, vinblastine and paclitaxel which is the exception of MRP7 unlike other MRPs (Huisman, Chhatta, van Tellingen, Beijnen, & Schinkel, 2005). It has been reported that MRP7 share only ~34 –36% of resemblance with other MRPs (Hopper et al., 2001). Like Multiple Resistant Protein 1, Multiple Resistant Protein 2, Multiple Resistant Protein 3 and Multiple Resistant Protein 6, Multiple Resistant Protein 7 also own an extra membrane spanning domain and so called “Long MRPs” and the others having no additional domain are called “Short MRPs” (Cole et al., 1992). Current investigation has reported that MRP7 also exhibit the efflux action like other ABC transporters (Z.-S. Chen et al., 2003; Kruh et al., 2007). Structurally MRP7 hold 22 exons and 21 introns which also shows its difference from other MRPs (Kao, Chang, Cheng, & Huang, 2003). High expression of MRP7 mRNA in the skin, colon, and testes, and in other tissues has been detected (Hopper et al., 2001). ABCC10 is analyzed as hydrophobic anion transporters which show resistance to different therapeutics agents including vinca alkaloids and taxanes (Hopper-Borge et al., 2004). MRP7 also allow resistance to antiviral agents such as Tenofovir and nucleoside-based agents Cytratine (Ara-C) and Gemcitabine (Pushpakom et al., 2011). Resistance mediated by MRP7 to paclitaxel, vincristine, docetaxel and vinblastine has been conveyed in an in-vitro study (Hopper-Borge et al., 2004). This study aims to provide the best 3D homology model of ABCC10. To identify the ligand site and its mechanism of ligand Binding Site. Homology modeling and Molecular docking against the ligand binding site may led to the identification novel ligands. Virtual screening of the model using Cambridge database was carried out to report some potential ligands against the ligand Binding Site of ABCC10. This study will provide an alternative to x-ray structure of ABCC10.
Number of Homology models of ABC transporter has been generated in so far, but the variation in the structure and transport properties of this diverse family of ABC transporters remain a major issue to reach to a final conclusion against these transporters. These models were generally constructed in the development of computational studies to address issues of multiple drug resistance in different diseases and primarily in cancer. The importance of structural elucidation of ABC transporters is a complex job. Due to large size of these transporters certain problems are posed. This study also focuses the generation of the best homology model of ABCC10. The overall process followed in this research is shown in the Figure 1 shown below.
2.1. Retrieving of Primary Sequence ABCC10
The primary sequence of ABCC10 (Accession No: Q5T3U5) of Homo Sapien was retrieved in FASTA format from the Universal Protein Resource (UniProt) (http://www.uniprot.org/).
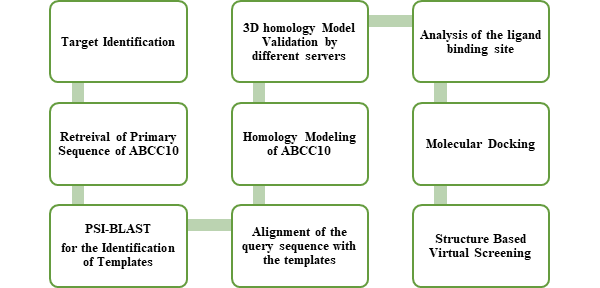
Figure 1: Showing the flow chart summary of the whole methodology followed in this thesis. Steps are followed as given.
2.2. Selection of Templates
A PSI-BLAST(Altschul et al., 1997) was carried out to search for templates sharing good homology with ABCC10 in the Protein Databank (PDB). PDB structures of highest homology with ABCC10 were retreived from Research Collaboratory for Structural Bioinformatics (RCSB) which were then used as tampletes against the query sequence of ABCC10. These proteins with PDB codes of tampletes with high sequnce identity were used for model building 4F4C, 2CBZ, 4C3Z, 3G5U, 3QF4, 4Q4J.
2.3. Alignment
An alignment of the retrieved template sequences were aligned on online T-coffee server (http://tcoffee.vital-it.ch/apps/tcoffee/result?rid=2c7281). The alignment of the query sequence (ABCC10) and other selected templates are shown in the Figure 2.

Figure 2: Showing the alignment of the query sequence with the selected templates used in the generation of final 3D homology model of ABCC10.
2.4. Trans-Membrane Domain & Intrinsic Diordered Regions Prediction
Topology of the proteins was predicted by using online Database Transporter Classification Database (http://www.tcdb.org/progs/TMS.php?anum=Q5T3U5&HIDE=1). PONDR (Predictor of Natural Disordered Regions) (http://www.pondr.com/cgi-bin/PONDR/pondr.cgi) server was used to predict the intrinsically disordered regions.
2.5. Analysis of Structural Properties
The structural properties of ABCC10 revealed that the chemical formula of ABCC10 is C7333H11703N1969O2046S45, molecular weight is 161628.07 and isoelectric focusing point is 7.07 using gene script (https://www.genscript.com/ssl-bin/site2/peptide_calculation.cgi). The phosphorylation sites for serine, threonine and tyrosine were predicted on NetPhos 2.0 Server (Blom, Gammeltoft, & Brunak, 1999).
2.6. Protein-Protein interaction network of ABCC10
Interaction of ABCC10 with other proteins and with ABC transporter family was analyzed. The interaction network revealed different kinds of interaction of ABCC10 with other proteins.
2.7. Homology Modeling of ABCC10
The generation of a good quality was aided with the generation of different metaservers and softwares including I-TASSER (Roy, Kucukural, & Zhang, 2010), PHYRE2 (Kelley, Mezulis, Yates, Wass, & Sternberg, 2015), SWISS-MODEL (Biasini et al., 2014), MODELER v 9.11 and MOE (Molecular Operating Environment). Each generated model was subjected to model validation tests but no good results were found. So, the final model was generated on Modeler by using the models obtained from different servers as tamplates. Number of homology models of ABCC10 was generated but due to low quality and just acceptable standard those models were dicarded using the models and selected templates in integration to generate a model which is accepted apreciably. These different models helped to prevent the formation of low quality model and built a model which was validated by different renowned model validation servers. The final model of ABCC10 was built on Modeler v 11 with the DOPE score of -158652.734375. Loops were automatically modelled using Modeler. Our analysis suggests that the use of multiple templates can provide excellent way to generate high quality homology model. Accordingly, we were able to find, suitable docking results of known inhibitors which can be considered as a fruit of high sequence identity. The loops regions were automatically refine by using Modeler v 9.11. the energy minimization of final full refine model was carried out by using an online energy minimization server YASARA (Krieger et al., 2009).
2.8. Validation of 3D Homology Model
Different online servers were used to validate the homology model of ABCC10 including Errat, ProQ, Rampage and Modeller DOPE score. The overall quality factor of this model showed this that the model is of high quality.
2.9. Ligand Binding Site Analysis
The ligand Binding Site of ABCC10 was analyzed by using the site finder tool of MOE and other online servers which confirm the ligand Binding Site s for ABCC10. The confirmation of the ligand Binding Site was also subjected to comparison with other ABC transporters both visually and analyzed the amino acids which showed the correct binding site of ABCC10. The docking of known ligands was carried out against the same given single binding site and then the results were analyzed for structure based virtual screening.
2.10. Ligand Molecules Selection and Molecular Docking
Very few inhibitors of ABCC10 have been documented so far. Initially the known ligands were docked against the ABCC10. The known ligands including Cyclosporine, Cytarabine, Daunorubicin, Docetaxel, Doxorubicin, Estradiol, Etoposide, Gemcitabine, Methotrexate, Paclitaxel, Sildenafil, Tenofovir, Verapamil and Vincristine (Shown in the Figure 3). The docking of known inhibitors for ABCC10 was carried out in Molecular Operating Environment (MOE 2014.0901) (Vilar, Cozza, & Moro, 2008). Initially only known inhibitors were docked against the ligand binding site. First the Molecular Database (.mdb) of in-vitro known 14 inhibitors was generated. Database of known ligands was generated, and energy minimization and protonation of each ligand was performed. All bonded and non-bonded interactions at a gradient of 0.05 Kcal/mol/Å. The database containing 14 known ligands was docked into the binding site using triangle matcher docking placement methodology. Thirty docking conformations were generated for each ligand and these conformations were ranked based on the free binding energies that were generated by London dG scoring function (Colotta et al., 2009; Magdziarz, Mazur, & Polanski, 2009).
3.1. Trans-Membrane Domain & Intrinsic Disordered Regions Prediction
Using TCDB and PONDR online servers to predict the transmembrane regions and the disordered regions in the structure of ABCC10. It was found that ABCC10 consists of 17 TMs regions while about 5.09% of the total sequence of ABCC10. The transmembrane are shown in table 1 while the disordered regions are shown in the Figure 4.
Table 1: Showing the transmembrane regions (Amino acids sequences, TM no along with their residues starting and ending) along with found in the structure of ABCC10.
|
Amino acids |
TM No |
Residues of TM |
|
LVLSALPHALLAVLSACYLGT |
TRANSMEMBRANE 1 |
32 – 52 |
|
LAASFLLSVFPLLDLLPVALP |
TRANSMEMBRANE 2 |
70 – 90 |
|
VLAGCVAAVAWISHSLALWVL |
TRANSMEMBRANE 3 |
102-122 |
|
LALALVALLPAPALVLTVLWH |
TRANSMEMBRANE 4 |
134-154 |
|
GTLLPPLLPGPMARLCLLILQ |
TRANSMEMBRANE 5 |
158-278 |
|
LALGLLKLVGTMLGFSGPLLLSLLVG |
TRANSMEMBRANE 6 |
286-311 |
|
GLLYALGLAGGAVLGAVLQ |
TRANSMEMBRANE 7 |
323–341 |
|
AGSFHEAWGLPLQLAITLYLL |
TRANSMEMBRANE 8 |
395–415 |
|
VGVAFVGGLILALLLVPVN |
TRANSMEMBRANE 9 |
419-437 |
|
AACVYLWAALPVVISIVIFITYVLMG |
TRANSMEMBRANE 10 |
504–529 |
|
VFTALALVRMLILPLNNFPWVINGLL |
TRANSMEMBRANE 11 |
537-562 |
|
AVGQGLALAILFSLLLMQATR |
TRANSMEMBRANE 12 |
879–899 |
|
VYATIAGVNSLCTLLRAVLFAAGTLQ |
TRANSMEMBRANE 13 |
970-995 |
|
SLPFILNILLANAAGLLGLLA |
TRANSMEMBRANE 14 |
1038–1058 |
|
SGLPWLLLLLPPLSIMYYH |
TRANSMEMBRANE 15 |
1062-1080 |
|
LQLMGAAVVSAIAGIALVQ |
TRANSMEMBRANE 16 |
1155–1173 |
|
LVGLSLSYALSLTGLLSGLVS |
TRANSMEMBRANE 17 |
1183-1203 |
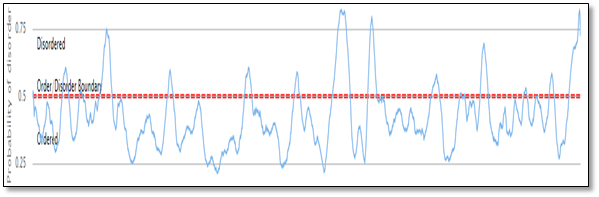
Figure 4: Prediction of the disordered regions in the sequence of ABCC10, which was found 5.09% of the total sequence of ABCC10.
3.2. Protein-Protein Interaction Network of ABCC10
The interaction of ABCC10 proteins with other proteins were predicted by using an online servers GeneMania (Warde-Farley et al., 2010) which showed the physical interaction, co-expression, pathways, co-localization, genetic interaction and domain-domain interaction was checked. This reveal that ABCC10 has domain-domain interaction with ABCB6 while with many others it has co-expression and also physical interaction. The interaction of ABCC10 was also checked with other ABC C transporters. The interaction of ABCC10 with other ABCs and other proteins family are shown in the Figure 5 & 6 respectively.
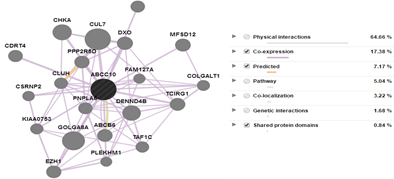
Figure 5: The interaction of ABCC10 with other proteins families. Their Physical, co-expression, pathway, localization, genetic and domain-domain interaction % has been shown in the figure.
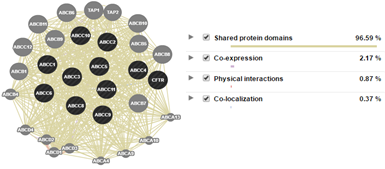
Figure 6: Interaction of ABCC10 with other ABC proteins. Their Physical, co-expression, pathway, localization, genetic and domain-domain interaction % has been shown in the figure.
Homology Modeling and Validiation of 3D Model of ABCC10:
The generation of homology model using multiple tamplets always favor a good output. Like other good homology models used multiple tampletes we have also generated our query protein homology model using multiple templates. The selected templates used in the construction of final homology model includes the MDR from C. elegans with 57% of sequence identity and 24% of coverage, Human MDR NBDomain 1 from Homo Sapien with 45% of sequence identity and 28% of coverage, Homo Sapien MRP1 with 42% of sequence identity and 32% of coverage, P-glycoprotein from Mus musculus with 35% of sequence identity and 50% of coverage, ABC transporter in its inward-facing conformation from Thermotoga maritima with 29% of sequence identity and 64% of coverage were used respectively to build the 3D homology model of ABBC10 (MRP7) using Modeler 9.11 v. Generally ≥30% of identity among query and templates are accepted for comparatively modeling (Forrest, Tang, & Honig, 2006). However, identity for membrane proteins ≥20% - 40% is widely accepted(Gao, 2009; Reddy, Vijayasarathy, Srinivas, Sastry, & Sastry, 2006). Different online servers were used to validate the homology model of ABCC10. The overall quality factor of this model showed this that the model is of high quality. Errat, ProQ, Rampage and Modeller DOPE score was used as model analyzer which confirm the quality of the model (Colovos & Yeates, 1993), (Lovell et al., 2003), (Wallner & Elofsson, 2003). The reliability of the model was verified by obtaining different results from these servers, which verify the 3D stability and validation of the model. The data showed below reflect the excellent quality of the model. After confirming on Errat the overall quality factor was 82.673%, which is considered as the best model. To obtain the Ramachandran Plot, Rampage server was used to plot the Ramachandran Plot of this model which showed that 88.9% (1324) amino acids lies in the favored region, 7.9% (118) amino acids plotted in allowed region while 3.2% (48) amino acids lies in the outlier region. ProQ (Protein Quality Predictor) (Maiti, Van Domselaar, Zhang, & Wishart, 2004) results showed that LGscore: 6.472 and MaxSub: 0.783 which clarify this that the model is built the best. Furthermore the model was verified by using TM Score (Template Modeling Score) which showed the overall TM score 0.718 as a TM score.0.5 indicates the best quality of the model(Xu & Zhang, 2010). Modeler DOPE score -158652.734375 also approved the model. The overall quality validation servers approved the model as the best and accurate model for Molecular Docking analysis. The final model of ABCC10 (Figure 7) and evaluation results obtained from Rampage and Errat are shown in the Figure 8 and 9 respectively.
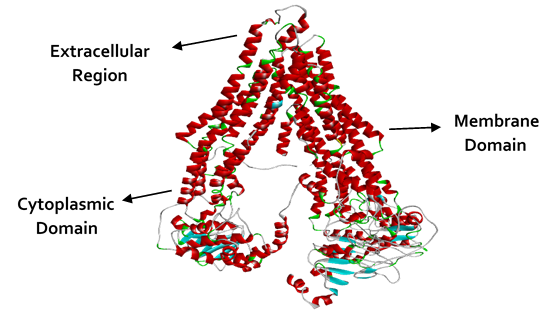
Figure 7: Ribbon illustration the final 3D homology model of ABCC10. The figure is also showing the extracellular regions, Membrane domain and the cytoplasmic domain of the modeled protein (ABCC10).
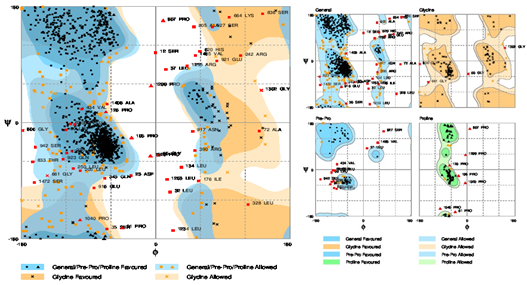
Figure 8: Ramachandran Plot showing the evaluation of the final model. The Plot showed 88.9% (1324) amino acids lies in the favored region, 7.9% (118) amino acids plotted in allowed region while 3.2% (48) amino acids lies in the outlier region.
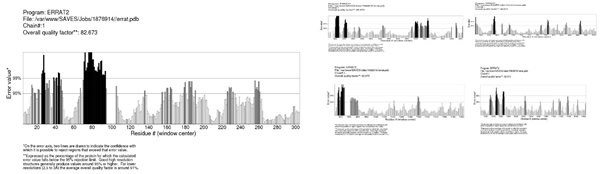
Figure 9: The Errat validation of the 3D homology Model of ABCC10. The overall quality factor showed by Errat was 82.673%. Due to large amino acids Sequence of the modeled protein the model is considered as very good with the overall quality showed.
3.3. Ligand Binding Site Analysis
The ligand Binding Site of ABCC10 (Figure 10) was analyzed by using the site finder tool of MOE, Discovery Studio Visualizer 4.5 Client and other online servers which confirm the ligand Binding Site s for ABCC10. The confirmation of the ligand Binding Site was also subjected to comparison with other ABC transporters both visually and analyzed the amino acids which showed the correct binding site of ABCC10. The binding site of ABCC10 comprising of residues are shown in the figure 9. The docking of known ligands was carried out against the same given single binding site and then the results were analyzed for structure based virtual screening.
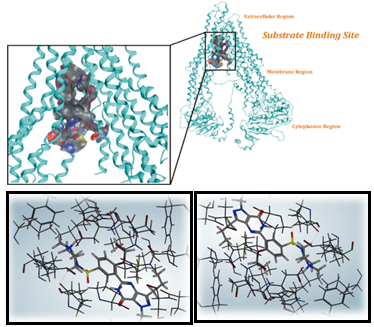
Figure 10: Illustration of the ligand Binding Site for ABCC10. The figure is showing the ligands docked against the given cavity.
3.4. Ligand Molecules Selection and Molecular Docking
For docking, the old drugs were repurposed to find the best one against the new target. The old drugs against ABCC10 were used include Cyclosporine, Cytarabine, Daunorubicin, Docetaxel, Doxorubicin, Estradiol, Etoposide, Gemcitabine, Methotrexate, Paclitaxel, Sildenafil, Tenofovir, Verapamil and Vincristine. The docking of these known ligands was followed by structure based virtual screening of the best affinity ligand. From the final list of docked conformation, the pose with good docking score was chosen for each ligand for the analysis. Gemcitabine was found to have good interaction with the receptor binding site. The docking score of the ligand is shown is in the table 2 and the best 2D interaction of the docked ligands along with their bonding pattern is shown in the Figure 11.
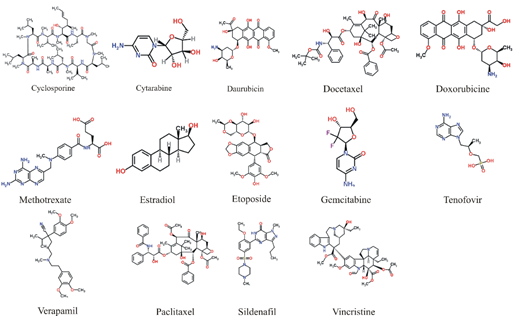
Figure 3: The figure is showing the 2D representation of the already known ligands used against ABCC10 for initial analysis.
Table 2: The docking score of the best ligands along with the number of Hydrogen bonds.
Conformation No |
Docking Score (Kcal/mol) |
No. of Hydrogen Bonds |
Amino Acid Interacted |
Bonds Distances (Å) |
|
1 |
-9.0922 |
4 |
Tyr226 |
3.12 Å, 3.06 Å |
|
32 |
-12.6801 |
2 |
Asp1037 |
2.90 Å, 2.85 Å |
|
96 |
-13.4099 |
1 |
Tyr226 |
3.25 Å |
|
126 |
-9.46163 |
2 |
Ser1029 |
2.64 Å, 2.76 Å |
|
178 |
-12.8231 |
1 |
Ser1030 |
3.07 Å |
|
196 |
-8.83444 |
2 |
Ser1030, Asp1037 |
3.04 Å, 3.06 Å |
|
238 |
-8.28571 |
4 |
Gln156, Ser219, Arg223 |
2.26 Å, 2.87 Å, 2.59 Å, 3.08 Å |
|
267 |
-12.5377 |
3 |
Lys438, Leu141, Asp1037 |
2.85 Å, 3.13 Å, 2.77 Å |
|
325 |
-16.8497 |
2 |
Ser1029 |
3.02 Å, 2.59 Å |
|
355 |
-15.4245 |
2 |
Asn437, Tyr226 |
1.96 Å, 2.06 Å |
|
385 |
-11.5758 |
2 |
Asn437, Tyr226 |
3.20 Å |
|
238 |
-8.28571 |
4 |
Gln156, Ser219, Arg223 |
2.26 Å, 2.87 Å, 2.59 Å, 3.08 Å |
3.5. Structure Based Virtual Screening
Gemcitabine docked pose within the binding site of ABCC10 was used to generate the pharmacophore model to find hits for virtual screening. A structure-based pharmacophore model was generated as shown in the Figure 12 by means of the MOE software and was used as a query to perform a virtual screening of Cambridge databases, followed by docking experiments. The interacted atoms were marked as essential feature to find hits in the available database. The features marked essentials includes three hydrogen bond Acceptor, one hydrogen bond Donor and Atom Q. The Cambridge database consists of 1, 72000 drugs. The pharmacophore was used as the three-dimensional query of a virtual screening approach. The top 5,151 compounds in terms of pharmacophore fit score were then submitted to docking studies by the MOE software. The screening of these compounds was carried out against the same binding site as used for the known ligands. For each ligand 30 conformations were allowed and the same docking protocol was followed as for known inhibitors. Of the 5,151 identified compounds a list of top scoring 50 drugs are given in the table 3.
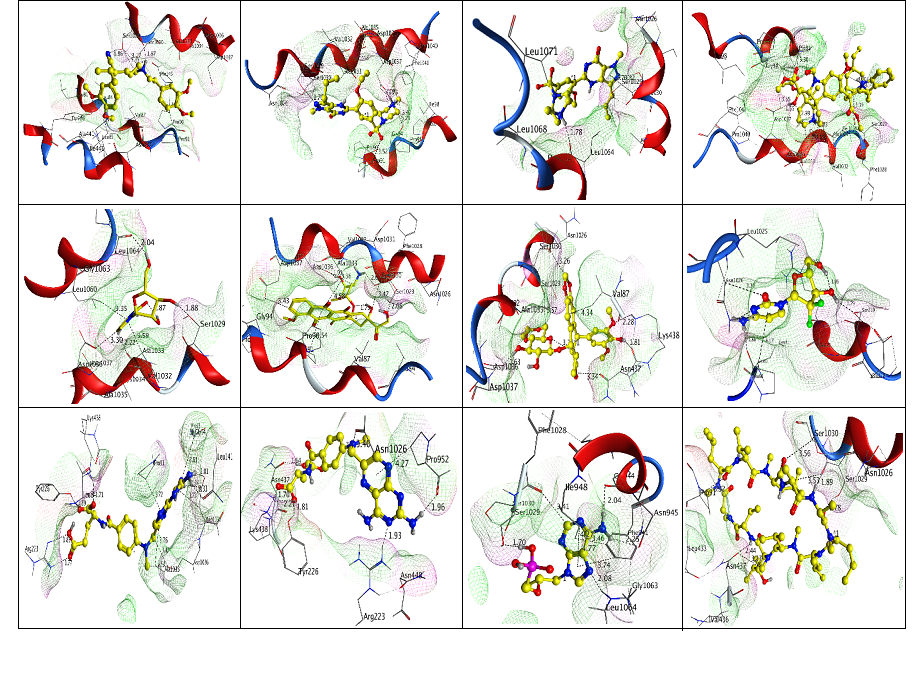
Figure 11: The depiction is showing the interaction of known ligands against with ABCC10 ligand Binding Site.
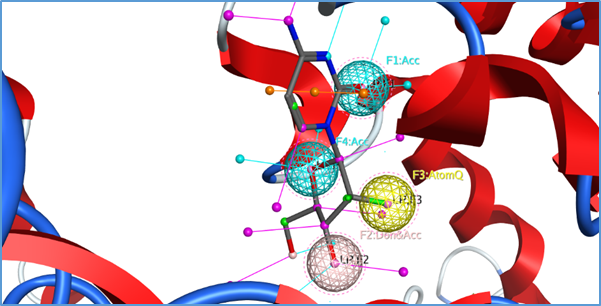
Figure 12: Showing the pharmacophore features of the interacted atoms marked as essential to search the Cambridge database to find the hits fitting the pharmacophore query.
Table 3: Showing the top 50 drugs of the 5,151 hits found to have activity against ABCC10. These ligands were selected on the basis of Docking score. The table is showing Index, rseq, mseq, Docking score, E_conf, E_place, E_score1 and E_score2.
|
Index |
rseq |
mseq |
Docking score |
E_conf |
E_place |
E_score1 |
E_score2 |
|
113143 |
1 |
3,808 |
-14.7538 |
1.6 |
-89.0189 |
-14.7538 |
-14.7538 |
|
133723 |
1 |
4,494 |
-14.372 |
1.2 |
-79.9003 |
-14.372 |
-14.372 |
|
103873 |
1 |
3,499 |
-14.2917 |
2 |
-72.8285 |
-14.2917 |
-14.2917 |
|
94813 |
1 |
3,197 |
-14.2505 |
0.605969 |
-71.6581 |
-14.2505 |
-14.2505 |
|
132793 |
1 |
4,463 |
-14.0407 |
2.2 |
-89.0752 |
-14.0407 |
-14.0407 |
|
35233 |
1 |
1,181 |
-13.987 |
2.6 |
-87.9322 |
-13.987 |
-13.987 |
|
86803 |
1 |
2,900 |
-13.9691 |
0.8 |
-97.0631 |
-13.9691 |
-13.9691 |
|
117373 |
1 |
3,949 |
-13.8565 |
1.8 |
-112.765 |
-13.8565 |
-13.8565 |
|
135583 |
1 |
4,556 |
-13.7359 |
0.836807 |
-55.5411 |
-13.7359 |
-13.7359 |
|
80113 |
1 |
2,677 |
-13.5761 |
3 |
-64.1843 |
-13.5761 |
-13.5761 |
|
54613 |
1 |
1,827 |
-13.5666 |
2.4 |
-104.457 |
-13.5666 |
-13.5666 |
|
64613 |
1 |
1,827 |
-13.5666 |
2.4 |
-104.457 |
-13.5666 |
-13.5666 |
|
36163 |
1 |
1,212 |
-13.5641 |
3.8 |
-101.943 |
-13.5641 |
-13.5641 |
|
54763 |
1 |
1,832 |
-13.5202 |
3.20001 |
-99.8712 |
-13.5202 |
-13.5202 |
|
64763 |
1 |
1,832 |
-13.5202 |
3.20001 |
-99.8712 |
-13.5202 |
-13.5202 |
|
103874 |
1 |
3,499 |
-13.5039 |
1.2 |
-71.3514 |
-13.5039 |
-13.5039 |
|
141163 |
1 |
4,742 |
-13.4864 |
2.8 |
-96.4193 |
-13.4864 |
-13.4864 |
|
123193 |
1 |
4,143 |
-13.46 |
3 |
-100.513 |
-13.46 |
-13.46 |
|
112273 |
1 |
3,779 |
-13.3521 |
2.6 |
-55.467 |
-13.3521 |
-13.3521 |
|
42623 |
1 |
1,427 |
-13.3271 |
1.2 |
-76.8594 |
-13.3271 |
-13.3271 |
|
76663 |
1 |
2,562 |
-13.3018 |
0.8 |
-66.8267 |
-13.3018 |
-13.3018 |
|
101533 |
1 |
3,421 |
-13.256 |
2.4 |
-55.6288 |
-13.256 |
-13.256 |
|
54493 |
1 |
1,823 |
-13.1564 |
2.2 |
-53.7726 |
-13.1564 |
-13.1564 |
|
64493 |
1 |
1,823 |
-13.1564 |
2.2 |
-53.7726 |
-13.1564 |
-13.1564 |
|
104953 |
1 |
3,535 |
-13.1154 |
3.46676 |
-128.247 |
-13.1154 |
-13.1154 |
|
142663 |
1 |
4,792 |
-13.079 |
2.41383 |
-65.3046 |
-13.079 |
-13.079 |
|
136573 |
1 |
4,589 |
-13.0505 |
2.8 |
-53.2578 |
-13.0505 |
-13.0505 |
|
22093 |
1 |
743 |
-13.0429 |
3.8 |
-81.3887 |
-13.0429 |
-13.0429 |
|
56803 |
1 |
1,900 |
-13.0424 |
2.8 |
-65.9371 |
-13.0424 |
-13.0424 |
|
66803 |
1 |
1,900 |
-13.0424 |
2.8 |
-65.9371 |
-13.0424 |
-13.0424 |
|
28843 |
1 |
968 |
-13.0387 |
1.8 |
-82.316 |
-13.0387 |
-13.0387 |
|
88063 |
1 |
2,942 |
-13.0302 |
2.6 |
-85.9968 |
-13.0302 |
-13.0302 |
|
105433 |
1 |
3,551 |
-13.0075 |
0.8 |
-93.1586 |
-13.0075 |
-13.0075 |
|
84913 |
1 |
2,837 |
-12.9948 |
1.45094 |
-53.1826 |
-12.9948 |
-12.9948 |
|
151153 |
1 |
5,075 |
-12.9755 |
2.4 |
-56.6204 |
-12.9755 |
-12.9755 |
|
83863 |
1 |
2,802 |
-12.9754 |
0 |
-54.1963 |
-12.9754 |
-12.9754 |
|
30373 |
1 |
1,019 |
-12.9519 |
3 |
-77.3477 |
-12.9519 |
-12.9519 |
|
98503 |
1 |
3,320 |
-12.9496 |
1 |
-72.6056 |
-12.9496 |
-12.9496 |
|
140953 |
1 |
4,735 |
-12.9493 |
2.6 |
-51.3369 |
-12.9493 |
-12.9493 |
|
117073 |
1 |
3,939 |
-12.9317 |
2.6 |
-83.237 |
-12.9317 |
-12.9317 |
|
12883 |
1 |
436 |
-12.9232 |
1.6 |
-71.6138 |
-12.9232 |
-12.9232 |
|
35023 |
1 |
1,174 |
-12.9024 |
0.6 |
-77.6521 |
-12.9024 |
-12.9024 |
|
148753 |
1 |
4,995 |
-12.8484 |
3.4 |
-100.583 |
-12.8484 |
-12.8484 |
|
128923 |
1 |
4,334 |
-12.834 |
2.4 |
-107.41 |
-12.834 |
-12.834 |
|
83293 |
1 |
2,783 |
-12.7941 |
4.04125 |
-71.6984 |
-12.7941 |
-12.7941 |
|
113144 |
1 |
3,808 |
-12.7938 |
0.4 |
-71.7851 |
-12.7938 |
-12.7938 |
|
23353 |
1 |
785 |
-12.7705 |
2.6 |
-77.6143 |
-12.7705 |
-12.7705 |
|
36313 |
1 |
1,217 |
-12.7454 |
0.6 |
-72.5222 |
-12.7454 |
-12.7454 |
|
59053 |
1 |
1,975 |
-12.745 |
0 |
-95.8198 |
-12.745 |
-12.745 |
Initially we reported the important features of ABCC10 which revealed that ABCC10 could be the best target for anti-cancerous drugs. Initial analysis suggested that ABCC10 is a long MRP 17 having transmembrane domains. We also reported the interaction of ABCC10 with other protein families and also other ABCs. It was reported that ABCC10 not only share homology with other proteins but also share co-expression, co-localization, domain-domain interaction and also physical interaction. We reported that ABCC10 in direct interaction with ABCB6 which could be a junction of great interest for drugs target. The generation of good homology model is always a tough task. The generation of 3D homology model of membrane proteins and large sequences is a laborious job. In this study the generated homology model of ABCC10 followed a very complex methodology. The initial templates selected for the generation of the model showed good homology and high coverage. We reported that the model was built best after using online servers which appreciable validate the model to be utilized for further analysis. The model obtained was similar and are in good agreement with ABC transporters. Analysis of the ligand binding site was more complex job than the model generation. The ligand Binding Site s were carefully analyzed using online tools and the site finder tools of MOE. By comparing the ligand Binding Site of our model with that of other models revealed that the ligand Binding Site of our modeled proteins is a common site like other MRPs. The residues of the ligand Binding Site were sharing homology with other MRPs and also visually the site can be analyzed. The residues analyzed for docking include Tyr226, Asp1037, Ser1029, Ser1030, Gln156, Ser219, Arg223, Lys438 and Leu141. We showed here that the revealed binding site is best for docking analysis. We also analyzed the in-vitro tested inhibitors against the selected binding site. Initially, we tested only 14 known ligands against the given site. The known ligands which we tested includes Cyclosporine, Cytarabine, Daunorubicin, Docetaxel, Doxorubicin, Estradiol, Etoposide, Gemcitabine, Methotrexate, Paclitaxel, Sildenafil, Tenofovir, Verapamil and Vincristine. The Index, rseq, mseq, Docking score, E_conf, E_place, E_score1 and E_score2 after docking revealed that all the in-vitro reported ligands against ABCC10 possess good activity. We also reported these compounds as best or not on the basis of number of hydrogen bonds formed only with the receptors but no other bonds. Our analysis suggested that conformers 1, 32, 61, 96, 126, 178, 196, 238, 267, 297, 325, 355 and 385 showed good interaction with the ligand Binding Site residues. Verapamil was found to have four interactions with the binding cavity of the receptor. The interacted residues we reported formed hydrogen bonds include Tyr226. The interacting residues Asp1037 was reported to form only 2 hydrogen bonds with Sildenafil. Estradiol was found in interaction with Tyr226 forming only single hydrogen bond. Further analysis revealed that Docetaxel showed interaction with Ser1029 with the bonding angles of 2.64 Å, and 2.76 Å respectively. Ser1030 was in interaction with Cytarabine. Our further analysis reported that Daunorubicin extended interaction with Ser1030, Asp1037 and showed good bonding angle.
In our further results we reported that Gemcitabine docked against the residues Tyr226, Asp1037, Ser1029, Ser1030, Gln156, Ser219, Arg223, Lys438 and Leu141 which resulted the formation of 4 hydrogen bonds with Gln156, Ser219, and Arg223 showing bonding angles of acceptable standard 2.26 Å, 2.87 Å, 2.59 Å, 3.08 Å. We also reported Methotrexate, Tenofovir, Cyclosporine, Paclitaxel and Cepharanthine interaction with the residues of binding site including Lys438, Leu141, Asp1037, Tyr226, Ser1029, Ser1030, Asp1037 and Ser219 respectively. Gemcitabine was reported as the best of all the ligands docked against the given site of ABCC10 receptor. Further, Gemcitabine was analyzed as essential features for pharmacophore search to carry out structure based virtual screening. The reported results indicated that out of the 1,72000 compounds only 5,151 compounds were having the same features like Gemcitabine. We reported that Gemcitabine showed hydrogen bond Acceptor, one hydrogen bond Donor and Atom Q as important features which were analyzed and found hits in the available database against the reported receptor. The features selected essential on the basis of bonding formed by Gemcitabine. We reported that these 5,151 compounds found fitted for the searched pharmacophore and could be test in-vitro to reported new drugs for cancer chemotherapy. Out of the 5,151 compounds found fit for structured based virtual screening, only top 50 compounds were reported. We suggest that in-vitro test of these compounds could lead to the evolution of new effective anti-cancerous drugs which could be effective against the multiple drugs resistant in cancer chemotherapy.
In this work, we have developed homology models of the ABCC10 transporter and assessed them in virtual screening for the identification of substrates. The models were generated by MOE, MODELLER, I-TASSER, EXPASY, PHYRE2. Energy minimization was carried out by using YASARA energy minimization server. The final model was built by combining all these models using MODELLER. The binding site was identified using MOE. The homology models were validated using different servers including Errat, Rampage, ProQ, and TM-Scoring. This approval was a confident way to dock known ligands against the binding pocket of ABCC10. Energy minimization, finding of disordered regions, secondary structure prediction and other evaluation was carried out to further confirm the validity of model. Binding site optimization, docking and Virtual screening was carried out. Initially the known ligands were docked which provide basis for the virtual screening other compounds. Gemcitabine and Methotrexate were found to have good docking score and found to have high hit (non-covalent) bonds). Finally, the Cambridge data base was used for virtual screening to identify further potential inhibitors which result in 5,151 were found to have good activity. This study provides a base for pharmacophore modeling and for molecular dynamic simulation which could further provide insight into the knowledge of ABC transporter family.
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., & Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic acids research, 25(17), 3389-3402. PMid:9254694
View Article PubMed/NCBIBiasini, M., Bienert, S., Waterhouse, A., Arnold, K., Studer, G., Schmidt, T., . . . Bordoli, L. (2014). SWISS-MODEL: modelling protein tertiary and quaternary structure using evolutionary information. Nucleic acids research, gku340. PMid:24782522
View Article PubMed/NCBIChen, J. J., Sun, Y. L., Tiwari, A. K., Xiao, Z. J., Sodani, K., Yang, D. H., . . . Chen, Z. S. (2012). PDE5 inhibitors, sildenafil and vardenafil, reverse multidrug resistance by inhibiting the efflux function of multidrug resistance protein 7 (ATP‐binding Cassette C10) transporter. Cancer science, 103(8), 1531-1537. PMid:22578167
View Article PubMed/NCBIChen, Z.-S., Hopper-Borge, E., Belinsky, M. G., Shchaveleva, I., Kotova, E., & Kruh, G. D. (2003). Characterization of the transport properties of human multidrug resistance protein 7 (MRP7, ABCC10). Molecular pharmacology, 63(2), 351-358. PMid:12527806
View Article PubMed/NCBIChen, Z. S., & Tiwari, A. K. (2011). Multidrug resistance proteins (MRPs/ABCCs) in cancer chemotherapy and genetic diseases. Febs j, 278(18), 3226-3245. doi:10.1111/j.1742-4658.2011.08235.x PMid:21740521
View Article PubMed/NCBIColovos, C., & Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein science: a publication of the Protein Society, 2(9), 1511. PMid:8401235
View Article PubMed/NCBIDean, M., & Annilo, T. (2005). Evolution of the ATP-binding cassette (ABC) transporter superfamily in vertebrates. Annu Rev Genomics Hum Genet, 6, 123-142. doi:10.1146/annurev.genom.6.080604.162122 PMid:16124856
View Article PubMed/NCBIDeeley, R. G., Westlake, C., & Cole, S. P. (2006). Transmembrane transport of endo-and xenobiotics by mammalian ATP-binding cassette multidrug resistance proteins. Physiological reviews, 86(3), 849-899. PMid:16816140
View Article PubMed/NCBIEckford, P. D., & Sharom, F. J. (2008). Interaction of the P-glycoprotein multidrug efflux pump with cholesterol: effects on ATPase activity, drug binding and transport. Biochemistry, 47(51), 13686-13698. doi:10.1021/bi801409r PMid:19049391
View Article PubMed/NCBIForrest, L. R., Tang, C. L., & Honig, B. (2006). On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophysical journal, 91(2), 508-517. PMid:16648166
View Article PubMed/NCBIGao, C. (2009). Computational studies on membrane protein structures and protein-ligand binding affinities. University of Rochester.
Gottesman, M. M., & Ambudkar, S. V. (2001). Overview: ABC transporters and human disease. J Bioenerg Biomembr, 33(6), 453-458. PMid:11804186
View Article PubMed/NCBIGottesman, M. M., Fojo, T., & Bates, S. E. (2002). Multidrug resistance in cancer: role of ATP-dependent transporters. Nature Reviews Cancer, 2(1), 48-58. PMid:11902585
View Article PubMed/NCBIHiggins, C. F. (2007). Multiple molecular mechanisms for multidrug resistance transporters. Nature, 446(7137), 749-757. doi:10.1038/nature05630 PMid:17429392
View Article PubMed/NCBIHopper-Borge, E., Chen, Z.-S., Shchaveleva, I., Belinsky, M. G., & Kruh, G. D. (2004). Analysis of the Drug Resistance Profile of Multidrug Resistance Protein 7 (ABCC10) Resistance to Docetaxel. Cancer research, 64(14), 4927-4930. PMid:15256465
View Article PubMed/NCBIHopper-Borge, E. A., Churchill, T., Paulose, C., Nicolas, E., Jacobs, J. D., Ngo, O., . . . Chen, Z.-S. (2011). Contribution of Abcc10 (Mrp7) to in vivo paclitaxel resistance as assessed in Abcc10−/− mice. Cancer research, 71(10), 3649-3657. PMid:21576088
View Article PubMed/NCBIHopper, E., Belinsky, M. G., Zeng, H., Tosolini, A., Testa, J. R., & Kruh, G. D. (2001). Analysis of the structure and expression pattern of MRP7 (ABCC10), a new member of the MRP subfamily. Cancer letters, 162(2), 181-191. 00646-7
View ArticleHuisman, M. T., Chhatta, A. A., van Tellingen, O., Beijnen, J. H., & Schinkel, A. H. (2005). MRP2 (ABCC2) transports taxanes and confers paclitaxel resistance and both processes are stimulated by probenecid. International Journal of Cancer, 116(5), 824-829. PMid:15849751
View Article PubMed/NCBIKelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., & Sternberg, M. J. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nature protocols, 10(6), 845-858. PMid:25950237
View Article PubMed/NCBIKimura, Y., Morita, S. Y., Matsuo, M., & Ueda, K. (2007). Mechanism of multidrug recognition by MDR1/ABCB1. Cancer Sci, 98(9), 1303-1310. doi:10.1111/j.1349-7006.2007.00538.x PMid:17608770
View Article PubMed/NCBIKrieger, E., Joo, K., Lee, J., Lee, J., Raman, S., Thompson, J., . . . Karplus, K. (2009). Improving physical realism, stereochemistry, and side‐chain accuracy in homology modeling: four approaches that performed well in CASP8. Proteins: Structure, Function, and Bioinformatics, 77(S9), 114-122. PMid:19768677
View Article PubMed/NCBIKruh, G. D., & Belinsky, M. G. (2003). The MRP family of drug efflux pumps. Oncogene, 22(47), 7537-7552. PMid:14576857
View Article PubMed/NCBIKruh, G. D., Guo, Y., Hopper-Borge, E., Belinsky, M. G., & Chen, Z. S. (2007). ABCC10, ABCC11, and ABCC12. Pflugers Arch, 453(5), 675-684. doi:10.1007/s00424-006-0114-1 PMid:16868766
View Article PubMed/NCBILovell, S. C., Davis, I. W., Arendall, W. B., de Bakker, P. I., Word, J. M., Prisant, M. G., . . . Richardson, D. C. (2003). Structure validation by Cα geometry: ϕ, ψ and Cβ deviation. Proteins: Structure, Function, and Bioinformatics, 50(3), 437-450. PMid:12557186
View Article PubMed/NCBIMaiti, R., Van Domselaar, G. H., Zhang, H., & Wishart, D. S. (2004). SuperPose: a simple server for sophisticated structural superposition. Nucleic acids research, 32(suppl 2), W590-W594. PMid:15215457
View Article PubMed/NCBIMalofeeva, E. V., Domanitskaya, N., Gudima, M., & Hopper-Borge, E. A. (2012). Modulation of the ATPase and transport activities of broad-acting multidrug resistance factor ABCC10 (MRP7). Cancer research, 72(24), 6457-6467. PMid:23087055
View Article PubMed/NCBINaramoto, H., Uematsu, T., Uchihashi, T., Doto, R., Matsuura, T., Usui, Y., . . . Yamaoka, M. (2007). Multidrug resistance-associated protein 7 expression is involved in cross-resistance to docetaxel in salivary gland adenocarcinoma cell lines. International journal of oncology, 30(2), 393-401.
Reddy, C. S., Vijayasarathy, K., Srinivas, E., Sastry, G. M., & Sastry, G. N. (2006). Homology modeling of membrane proteins: a critical assessment. Computational biology and chemistry, 30(2), 120-126. PMid:16540373
View Article PubMed/NCBIRoy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols, 5(4), 725-738. PMid:20360767
View Article PubMed/NCBISchinkel, A. H., & Jonker, J. W. (2003). Mammalian drug efflux transporters of the ATP binding cassette (ABC) family: an overview. Adv Drug Deliv Rev, 55(1), 3-29. 00169-2
View ArticleShen, T., Kuang, Y.-H., Ashby Jr, C. R., Lei, Y., Chen, A., Zhou, Y., . . . Ouyang, J. (2009). Imatinib and nilotinib reverse multidrug resistance in cancer cells by inhibiting the efflux activity of the MRP7 (ABCC10). PloS one, 4(10), e7520. PMid:19841739
View Article PubMed/NCBITakayanagi, S.-i., Kataoka, T., Ohara, O., Oishi, M., Kuo, M. T., & Ishikawa, T. (2004). Human ATP-binding cassette transporter ABCC10: expression profile and p53-dependent upregulation. Journal of experimental therapeutics & oncology, 4(3).
Wallner, B., & Elofsson, A. (2003). Can correct protein models be identified? Protein Science, 12(5), 1073-1086. PMid:12717029
View Article PubMed/NCBIWarde-Farley, D., Donaldson, S. L., Comes, O., Zuberi, K., Badrawi, R., Chao, P., . . . Lopes, C. T. (2010). The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic acids research, 38(suppl 2), W214-W220. PMid:20576703
View Article PubMed/NCBIWooden, S. L., Kalb, S. R., Cotter, R. J., & Soloski, M. J. (2005). Cutting Edge: HLA-E Binds a Peptide Derived from the ATP-Binding Cassette Transporter Multidrug Resistance-Associated Protein 7 aSnd Inhibits NK Cell-Mediated Lysis. The Journal of Immunology, 175(3), 1383-1387. PMid:16034073
View Article PubMed/NCBIXu, J., & Zhang, Y. (2010). How significant is a protein structure similarity with TM-score= 0.5? Bioinformatics, 26(7), 889-895. PMid:20164152
View Article PubMed/NCBI