Zhang Rui
Email: zhangrui168@gdou.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 5 ISSUE: 1
Page No: 35-43
Zhang Rui
Email: zhangrui168@gdou.edu.cn
ZhangRui1,2*, ChenWei-Ling2, HuangXing-Yun2, SunMei-Rong4, ZhangHong-Lian1,2 , Cheng Weng Jie1, WangRong-Lian1,3, DuHui1,2, LiaoYan1,2
1Shenzhen Institute of Guangdong Ocean University, Shenzhen, Guangdong518120, P. R. China;
2 College of Food Science and Technology, Modern Biochemistry Experimental Center, Guangdong Ocean University, Zhanjiang, Guangdong 518088, P. R. China;
3 College of Agriculture, Guangdong Ocean University, Zhanjiang, Guangdong 524088, P. R. Chi- na;
4 Shenzhen GuiBao technology co. LTD
Zhang Rui, New methods of mold control in animal specimens(2020)Journal of Earth Sciences & Environmental Studies 5(1) pp:35-43
Mold spores and hyphae infection is a serious problem and can lead to damage or loss of natural history specimens. Animal specimens play an important role in cultural exchange, science popularization, scientific research and economic window, but the preparation and preservation technology system of biological specimens is relatively unsafe and inefficient. Infection by Mold spores and hyphae is not only an acute problem that may cause damage or loss of specimens,but alsois very harmful to human beings health and polluted to environment, It is one of the principal element that restricts the development of the natural history museums. In this paper, identified the mold species of animal specimens by PCR with ITS primers, bio-microscopic observation, sequencing and phylogenetic tree analysis. The results showed the mold of animal specimens mainly belong to Aspergillus and Neurospora. than screened environmental friendly fungicides, the results show that both chitosan and nano silver can effectively inhibit the Aspergillus and Neurospora of animal specimens , the study provides a theoretical basis for the study of the new specimen fungicides and the environmental safety of the natural history museum
Key words: Animal specimens, mold identification, phylogenetic tree analysis, Mold fungicides, chitosan; nano-silver
1.1 Current situation
At present, there are many poisonous reagents used in preparation of animal specimens. Such as arsenic trioxide ( As2O3 ), Camphor, Phenol, Oxymethylene, and so on. It is harmful to people’s health and the environment of exhibition, such as conservation workers, and the general public.Mold is the most important role to damage the animal specimens [Fig1]. . Exposure to hot and humid environments has damaged specimens. This environment has been especially hard on animals that have been taxidermied because of they have external skin, fur, internal fat, protein, and connective tissue , etc.,. Infection by mold spores and hyphae is an acute problem to animal specimens, which have been associated with many human beings serious health problems and environmental safety problems. It would also lead to great loss of specimens in natural history collections. Because animal specimens are more precious than any other forms of biological specimens such as plants and other specimens. They are very expensive, so it would be a huge economic lost if they were damaged. South China has been hot and humid climate for about 8-10 months. In Guangdong province, the annual average relative humidity is 65-85%. Especially during the mold rains season, the relative humidity often reached more than 90%, sometimes even was 100%.

Figure1. There are many poisonous reagents used in preparation of animal specimens.
At present, there are many reports on the ancient cultural relics such as paper, ancient lacquer painting, silk cotton fabric, leather, ivory, bamboo, jade, pottery and porcelain, glass [1-4]. Recently there have been a few reports on the mold that infected animal specimens,.
So, there are great challenges and difficulties in the most important techniques for sample collection, preparation and storage of animal specimens.
1.2 Mold Damage: The harmfulness of animal specimens Mold to Human beings
The incidence and prevalence of serious mycoses continues to be a public health problem.
Thousands of species of microscopic MOLD are common in the environment, these mold feed on dead organic matter, including natural history specimens. The species involved Aspergillus, Penicillium, Geotrichum, Alternaria, Cladosporium, Mucor, Rhizopus, Trichoderma, Fusarium which are associated mostly with allergic response of different types. At first, mold is harmful to human beings. Microbiological contamination with mold, It not only can pose a significant health hazard to those working in archives or museums[5], but also could lead to many serious illness. The nose and paranasal sinuses almost always harbor numerous mold species. Amongst the species recovered, many have the potential to produce mycotoxins including Aspergillus (flavus, niger, fumigatus, versicolor), Chaetomium, Fusarium, Penicillium and Trichoderma. This group also found “mold elements (hyphae, destroyed hyphae, conidiae and spores)”. Mold and eosinophilic mucin are the markers of sinus involvement in the CRS patients[6]. Chronic Rhinosinusitis (CRS) , A primary concern was that exposure to high concentrations of Aspergillus spores may cause respiratory illness, particularly in individuals that are immunocompromised, it also led to endocrinopathy with growth hormone deficiency; Dennis-Robertson syndrome[7-9]. Molds and mycotoxins cause neurobehavioral and pulmonary impairment as well as adverse health effects especially with allergies and asthma. [10-19]. So, it is very important to Characterize and analyze the mold of animal specimens.
1.3 Antibacterial mechanism of new environment-friendly anti-mildew agent
1.3.1 Antibacterial principle of chitosan
Chitosan is a kind of anti-microbial substance, which ACTS in the following ways: damage cell wall, change cell permeability, change protein and nucleic acid molecules, inhibit the action of enzymes, as an anti-metabolite, inhibit nucleic acid synthesis.About the antibacterial mechanism of chitosan and its derivatives, from the point of the current study results, mainly include the following: (1) several possible molecular weight is less than 5000 kda chitosan can through the cell membrane, small molecular chitosan into the microbial cells, and cells negatively charged material (mainly proteins and nucleic acids), make the normal physiological function of the cell (such as DNA replication and protein synthesis, etc.), killing microorganisms.(2) chitosan of macromolecules adsorbed on the surface of microbial cells, forming a layer of molecular membrane, preventing the transport of nutrients to cells, thus playing a bactericidal and bacteriostatic role.(3) the interaction between the positive charge of chitosan and the negative charge on the surface of the microbial cell membrane changed the permeability of the microbial cell membrane and caused the microbial cell death.(4) chitosan, as a chelating agent, selectively chelates metal ions that play a key role in microbial growth, thus inhibiting microbial growth and toxin production, which may be one of the reasons for the preservative and preservation function of chitosan.(5) chitosan can activate chitinase activity of the microorganism itself. When the concentration of chitosan is high enough, chitinase of the microorganism is overexpressed, leading to chitin degradation of its own cell wall, thus damaging the cell wall.
1.3.2 Antibacterial principle of nano silver
Nano silver is to achieve the particle size of nano silver powder, particle size is less than 100nm, generally between 25 nm and 50nm.The silver nanoparticles directly enter the bacteria and combine with the oxygen metabolism enzyme (-sh) to inactivate the enzyme and block the unique mechanism of respiratory metabolism that asphyxiates and kills the bacteria, which can kill most bacteria, fungi, molds, spores and other microorganisms it comes into contact with.Through the experiment, it kills more than 650 kinds of bacteria such as graminella, graminella, fungus, gonococci, chlamydia trachomatis and so on in a few minutes, and has broad spectrum sterilization without any drug resistance.The performance of silver nanoparticles is directly related to their particle size. The smaller the particle size is, the higher the valence state is and the stronger the bactericidal performance is. The silver nanoparticles with the size of 10nm can quickly and directly kill bacteria and make them lose the ability to reproduce.Silver nanoparticles have super permeability, can quickly infiltrate under the skin 2nm sterilization, common bacteria, stubborn bacteria, drug-resistant bacteria and fungi caused by the deeper tissue infection have a good permanent sterilization effect.Nano silver non-toxic, tasteless, pollution-free, no skin irritation, no carcinogenicity, no harm to human body.
Nano-silver has been widely used in medical and sanitary products, but no literature has been reported on its use in specimen preparation and preservation.With the development of nano-silver preparation technology, the cost of nano-silver has been greatly reduced, so the price has been reduced to a reasonable level, it will be possible to use it in specimen preparation and preservation.
2.1 Experimental materials
The mold samples were collected from animal specimens of Shenzhen museum during December 2015-March 2016. These specimens include porcupine, birds, turtles, antelope and deer. Because of the mould rainy day, there are plenty of mold exposed on them. (Figure 2)
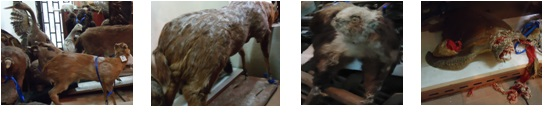
Figure2. The mold samples was collected from animal specimens.
The chemical reagent was purchased from guangzhou xiangbo biotechnology co., LTD.
2.2 Methods
2.2.1 Mold cultured and isolated and microscopes observation
PDA medium was used to culture and isolate the collected mold. Identification of the mold was confirmed by culturing swabs from infected specimens and examining mold hyphae with both light bio-microscopes .
The swabs were immersed and homogenized in a sterile physiological solution and serial dilutions were made. Each dilution was inoculated (0.1 ml) on MEA. After 2 days of cultivation at 25˚C, identification of the mold was performed. After 3 days of fostering, seemingly different mold colony grew on 12 plates, A small sample from the middle of the colony was dug and transferred to new plate to culture the different colonies. Strains that were previous activated had a small amount of sterile water with the pipettor added. The mold was then extracted from the solution and inoculated on a slant medium. This was repeated.
Then slightly digged a small piece of colony from the center of the same kind. Mold were transferred to a new plate one by one until separating into single and stable colonies plates. Take strains previously activated and kept in the AGAR slant tube, added some amount (bacteriostatic ring according to the experimental plate number) of sterile water with the pipettor on the clean work platform, then moved the pipettor under the liquid level, absorbed a little liquid and gently hit out, beat the mold on the inclined medium, let more mold into the suspension, make of the mold suspension.
The mold was observed through bio-microscopic observation. A small amount mold hyphae was dropped onto the water slide to make a suspension. At this point, the class slide was turned 45 degrees slowly to cover the mold to make observations of the bubbles. The slide was put on the staged and to observe the structure of the mold. Computer software was then used to characterize the mold.
Cultural and micromorphological characteristics of mold colonies observation and identification was performed through using identification ways.
The collection consisted of deteriorated animal specimens, including 12 species of animal specimens, 1. Porcupine 2.wood, 3.bird, 4.turtle, 5.antelope, 6 deer1. 7. Deer2, 8.wale. 9.chimpanzee, 10. Raja porosa
2.2.2 PCR detection
The isolated single colonies and ITS primers were used to do PCR(S1000 Thermal Cycler BioRad, USA). Primers were ITS1 and ITS4. The mold DNA was amplified using the highly conserved fungal rRNA gene primers (ITS1F and ITS4) as White and Gardes previously described [20-22] . The sequence of ITS1F and ITS4 are
ITS1F 5’-CTTGGTCATTTAGAGGAAGTAA-3’ ,
ITS4 5’-TCCTCCGCTTATTGATATGC-3’
PCR Reaction conditions was 94℃ predenatured for 5 min, 98℃ denatured for 30s, 58℃ annealed for 30s, 68℃ extended for 2-3 min. After the total 27 cycles, They were detect by agarose gel electrophoresis.
2.2.3 Sequencing and phylogenetic tree analysis
The isolated single colonies were sent to ShangHai ShengGong Company, Ltd to be sequenced. And the results were analyzed. Distance-matrix sequence alignment methods used Clustal W. Maximum likelihood (ML) phylogenetic tree for the fungal Operational Taxonomic Units (OTUs) constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. These sequences resulted in 8 fungal Operational Taxonomic Units (OTUs) with ITS and 19 OTUs with 18S rDNA primer sets respectively by taking into account the 2% sequence divergence cut-off for species delineation. Topology was built using Mega v.5.03 from a ClustalW 1.83 alignment. Numbers below branches indicate bootstrap values (>50%) from 1,000 replicates. New sequence types are marked with triangle. B: Proportional distribution of different fungal taxa in the clone library constructed with fungal specific primer set ITS1F/ITS4
2.4 Mold suppression experiment
Configured a certain concentration of chitosan and nano silver solution, At first coated the Purified mould liquid on a dish, then coated different concentrations of Mould inhibitors, formaldehyde as a control, and then cultivate 24 to 48 hours (decision) according to the bacterial growth status, microscope to observe the antibacterial effect.
Identification of the mold was confirmed by culturing swabs from infected specimens and examining mold hyphae both light bio-microscopes. Mold microscopic observation of 12 mold infected species were identified
The examined animal specimens in Shen Zhen museum showed clear signs of biodeterioration. Superficial colonies of mold were clearly visible and abundant., A total of 12 mold species were identified from all analyzed samples with 12 taxa identified (Figure. 3).
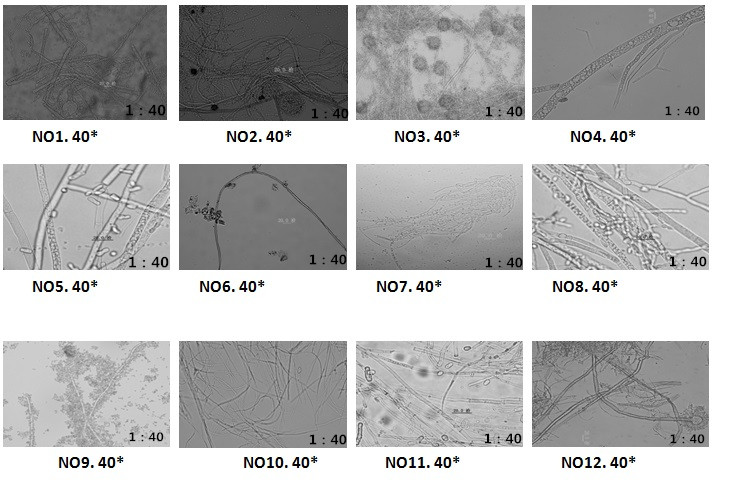
Figure3. Mold hyphae microscopic observation that isolated from 12 mold that infected animal specimens.
The mold hyphae was detected used 40 fold light bio-microscopes, It need to improve the fold of bio-microscopes, It would have better observation results.
3.2 Single colony-PCR detection
Because the mold collected from the museum was mixed mold samples, they were still unable to determine the type and the single degree, so PCR was used for species identification. The isolated single colonies were detected by PCR primers ITS1 and ITS4. The results showed that the DNA bands were right (Figure 4).

Figure4. Colony-PCR, ITS primer be used to identify of mold samples, 1-12 mold samples collected from 12 spieces . 1. Porcupine 2.wood, 3.bird, 4.turtle, 5.antelope, 6 deer1, 7. Deer2, 8.wall. 9.chimpanzee, 10. Raja porosa
M: Marker, Del2000, 1-12 was the mold samples PCR that was collected from animal specimens.
The above figure 5 was the electrophoresis stripes after the PCR amplification of 1 ~ 12 strains, could see the bright and clear 12 stripes from the figure, colony PCR experiment successfully copied the genetic material of 12 strains.
Twelve animal species mold samples DNA were amplified through colony PCR amplification. Then it were done the species identification. The results proved that the colony PCR experiments could be successfully used for species identification.
Mold genetic materials were amplified in colony PCR expansion test and species identification was performed. The results proved that the colony PCR experiments could be successfully used for species identification.
The figure 5 showed the electrophoresis bands after the PCR amplification of 1 ~ 12 strains, It could be seen the bright and clear 12 bands from the figure, Colony PCR experiment successfully copied the genetic material of 12 strains.
Colony PCR constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. But the general primers sometimes had some limited, It would have better PCR results after screening advanced specific primers[23].
3.3 Sequenced and analyzed, phylogenetic tree analysis
The single isolated colony was sent to ShangHai ShengGong Company, Ltd to be sequenced. The results were analyzed. Distance-matrix sequence alignment methods used Clustal W (Figure 5). The results showed the mold from animal specimens mainly belong to Aspergillus and Neurospora. The results were benefit for different novel mold inhibitor screening according to the different types of mold that molecular species identification.
Phylogenetic tree for the fungal Operational Taxonomic Units (OTUs) constructed based on fungal ITS gene sequences obtained using fungal specific primer set ITS1F/ITS4. These sequences resulted just in fungal Operational Taxonomic Units (OTUs) with ITS and 19 OTUs with 18S rDNA primers that with different fungal taxa in the clone library constructed with fungal specific primer set ITS1F/ITS4. But fungal strains used for specificity test of new 28S rDNA primers would be more significant results[24-26].
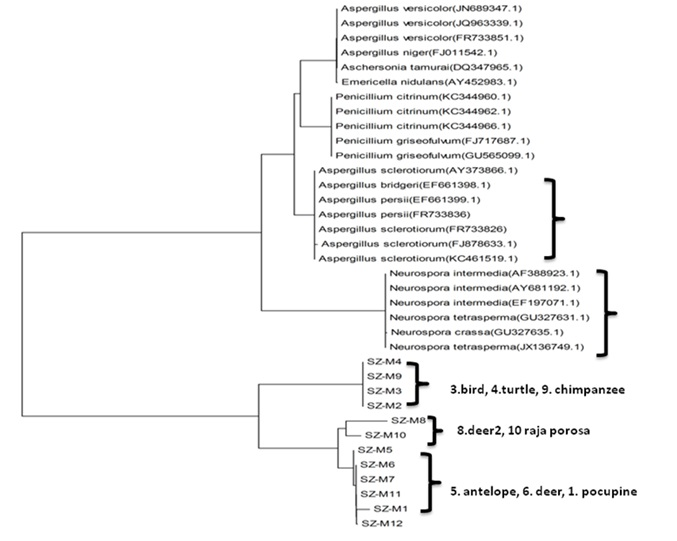
Figure5. Mold genetic Sequenced and analysed, phylogenetic tree analysis
The results showed the prevailing mold species isolated from animal specimens was Aspergillus niger Tiegh (62.5%), followed by Neurospora crassa Shear & B.O. Dodge (25%)
The signal colony that isolated sent to ShangHai ShengGong Company, Ltd to be sequenced. And the results was analysied, Distance-matrix sequence alignment methods was used Clustal W.
3.4 The effect of Mold fungicides
It Can be show from figure 1 with fluorescent microscope under 40-x , the sporangium,spore , hyphae and the mycelia of the molds, the significant results showed that both chitosan and nano silver can effectively inhibit the Aspergillus, It has a good effect on mold inhibition(Figure 6).
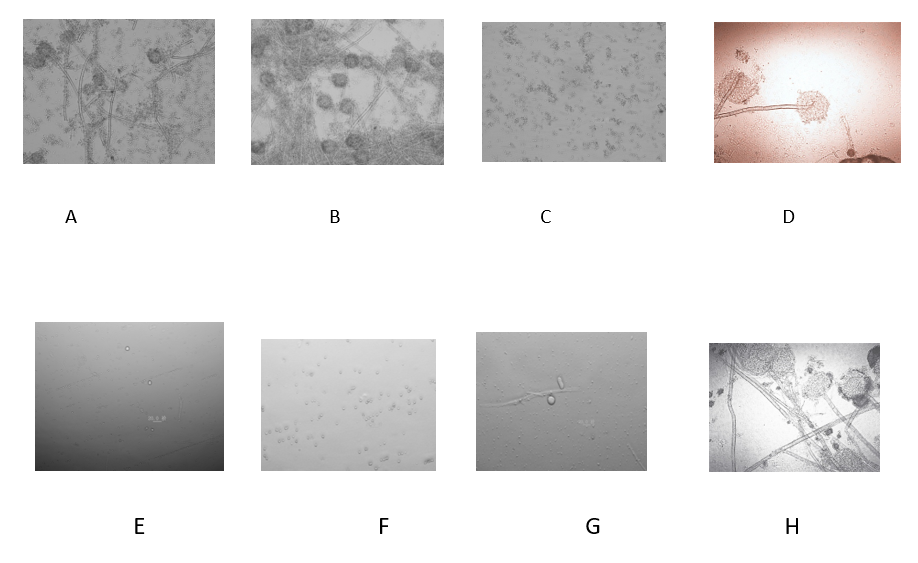
Figure6. Microscopic observation of bacteriostatic effect after cultured 30 hours
A. Chitosan 10ppm, B. Nano silver 10ppm, C. Chitosan+Nano silver, respectively10ppm, D. Control
E. Chitosan 100ppm, F. Nano silver 100ppm, G. Chitosan+Nano silver, respectively100ppm, H. Control
Identifying the mold species from animal specimens, by PCR with ITS primer, bio-microscopic observation and sequencing and phylogenetic tree analysis, the results showed the mold from animal specimens mainly belong to Aspergillus and Neurospora. This study established the foundations of controlling and restoring the mold that infected animal specimens and guided a new methodology of preparation and environmental friendly exhibition for animal specimens. It was benefit for different novel mold inhibitor screening according to the different types of mold that molecular species identification. And both chitosan and nano silver can effectively inhibit the Aspergillus, It has a good effect on mold inhibition
This research was supported by Industrial develop- ment special fund project of Shenzhen Dapeng New District (KY20180204)
Guangdong provincial innovation and entrepreneur- ship training program for college students (CXXL2017039)
Kolomodin-Hedman B, Blomquist G, Sikstrom E(1986) : Mould exposure in museum personnel. Int Arch Occup Environ Health, 57(4), 321-323. PMid:3710604
View Article PubMed/NCBIKrake AM, Worthington KA, Wallingford KM, Martinez KF(1999): Evaluation of Microbiological Contamination in a Museum. App Occup Environ Hyg , 14(8), 499-509.
View ArticleJeyaprakasam, NK ,Razak, MFA , Ahmad, NAB, Santhanam, J(2016) : Determining the Pathogenic Potential of Non-sporulating Molds Isolated from Cutaneous Specimens, 181(5-6): 397-403 PMid:26847667
View Article PubMed/NCBIKrysińska-Traczyk E(1994): Contamination of archives by filamentous fungi and their evaluation for potential pathogenicity. Med Pr, 45(6), 495-500 .
Lugauskas A, Krikstaponis A: Microscopic Fungi Found in the Libraries of Vilnius and Factors Affecting their Development. Indoor Built Environ 2004, 13 (3), 169-182.
View ArticleKatarzyna Zielińska-Jank , Anna Kozajda , Małgor(2008), Microbiological contamination with mould in work environment in libraries and archive storage facilities. Ann Agric Environ Med, 15, 71-78.
Brewer, J.H.; Thrasher, J.D.; Straus, D.C.; Madison, R.A.; Hooper, D(2013). Detection of mycotoxins in patients with chronic fatigue syndrome. Toxins, 5, 605-617. PMid:23580077
View Article PubMed/NCBIDennis, D.P(2003). Chronic defective T-cells responding to superantigens, treated by reduction of fungi in the nose and air. Arch. Environ. Health, 58, 433-451. PMid:15143856
View Article PubMed/NCBIDennis, D.P.; Roberson, D.; Curtis, L.; (2009) Fungal exposure endocrinopathy with growth hormone deficiency; Dennis-Robertson syndrome. Toxicol. Ind. Health, 25, 669-680. PMid:19808744
View Article PubMed/NCBIRea, W.J.; Didriksen, N.; Simon, T.R, Griffiths, G(2003). Effects of toxic exposure to mold and mycotoxins in building-related illnesses. Arch. Environ. Health, 58, 399-405. PMid:15143852
View Article PubMed/NCBICampbell, A.; Thrasher, J.D.; Gray, M.R.; Vojdani, A(2004). Mold and mycotoxins: Effects on the neurological and immune systems in humans. Adv. Appl. Microbiol, 55, 375-398. 55015-3
View ArticleGray, M.R.; Thrasher, J.D.; Crago, R (2003), Mixed mold mycotoxicosis: Immunological changes in humans following exposure to water damaged buildings. Arch. Environ. Health, 58, 410-420. PMid:15143854
View Article PubMed/NCBIKilburn, K.H. Neurobehavioral and pulmonary impairment in 105 adults with indoor exposure to molds compared to 100 exposed to chemicals. Toxicol. Ind. Health 2009, 35, 681-692. PMid:19793776
View Article PubMed/NCBIC.O. Mohan,C.N. Ravishankar,K.V. Lalitha,T.K. Srinivasa Gopal(2011). Effect of chitosan edible coating on the quality of double filleted Indian oil sardine ( Sardinella longiceps ) during chilled storage[J]. Food Hydrocolloids,26(1):21-29
View ArticleJ. Gómez-Estaca,A. López de Lacey,M.E. López Caballero,M.C. Gómez-Guillén,P. Montero. Biode gradable gelatin-chitosan films incorporated with essential oils as antimicrobial agents for fish preser vation[J]. Food Microbiology,2010,27(7). PMid:20688230
View Article PubMed/NCBIFisk, W.J.; Eliseeva, E.A.; Mendell, M.J(2010). Association of residential dampness and mold with respiratory tract infections and bronchitis: A meta-analysis. Environ. Health, 9, 72-84. PMid:21078183
View Article PubMed/NCBIEl-Morsy, S.M.; Khafagy, Y.W.; El-Naggar, M.M.; Beih, A.A. (2010). Allergic fungal rhinosinusitis: Detection of fungal DNA in sinus aspirate using polymerase chain reaction. J. Layrngol. Otol. 2010, 124, 152-160. PMid:19968888
View Article PubMed/NCBIVennewald, I., Wollina, U(2005).. Cutaneous infections due to opportunistic molds: Uncommon presentations ,Clinics in Dermatology, 23 (6):565-571 PMid:16325064
View Article PubMed/NCBILionakis, M.S., Kontoyiannis, D.P(2004). Fusarium Infections in Critically Ill Patients,Seminars in Respiratory and Critical Care Medicine , 25 (2):159-169 PMid:16088459
View Article PubMed/NCBIBiobanking in microbiology (2005) : From sample collection to epidemiology, diagnosis and research , Paolo De Paoli, EMS Microbiology Reviews, 29, 897-910 PMid:16219511
View Article PubMed/NCBILjaljevic-Grbic, M.; Stupar, M.; Vukojevic, J.; Maricic, I (2013). Bungur, N. Molds in museum environments: Biodeterioration of art photographs and wooden sculptures. Archives of Biological Sciences, 65, 955-962.
View ArticleBush and Portnoy, (2001).The presence of fungal propagules in indoor air causes adverse health effects, especially allergies and asthma. 2, 59-64.
White, T.J., Bruns, T.D., Lee, S.B., Taylor, J.W., (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In:Innis,M.A., Gelfand, D.H., Sninsky, J.J.,White, T.J. (Eds.), PCR Protocols-a Guide to Methods and Applications. Academic Press, San Diego, CA. 315-322. PMid:1696192
View Article PubMed/NCBIGardes, M., Bruns, T.D., (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113-118. PMid:8180733
View Article PubMed/NCBILai X, Cao L, Tan H, Fang S, Huang Y, Zhou S (2007) Fungal communities from methane hydrate-bearing deep-sea marine sediments in South China Sea. ISME J. 1, 756-762 PMid:18059498
View Article PubMed/NCBIDaniel K. M , Jorge M. V , (2007). Use of the ITS primers, ITS1F and ITS4, to characterize fungal abundance and diversity in mixed-template samples by qPCR and length heterogeneity analysis.Journal of Microbiological Methods 71, 7-14 PMid:17683818
View Article PubMed/NCBIPurnima S, Chandralata R, Pankaj V, Yogesh S, (2012). Assessment of fungal diversity in deep-sea sediments by multiple primer approach World J Microbiol Biotechnol 28:659-667 PMid:22806861
View Article PubMed/NCBIRebecca C Mueller1, Laverne G G, Donald R. Z,(2016). Assembly of Active Bacterial and Fungal Communities Along a Natural Environmental Gradient, Microb Ecol 71:57-67. PMid:26280745
View Article PubMed/NCBIDirk Krüger D K, Christiane F R, DanielT W,( 2012), Diversity Measures in Environmental Sequences Are Highly Dependent on Alignment Quality-Data from ITS and New LSU Primers Targeting Basidiomycetes, Plos one, 20, 5-25.
Sandra M. Velasco-C, Elkin Aguirre-R , Jenny Johana G-F (2019) Saving DNA from museum specimens: The success of DNA mini-barcodes in haplotype reconstruction in the genus Anastrepha (Diptera: Tephritidae), Journal of Advanced Research 16 :123-134 PMid:30899595
View Article PubMed/NCBI