Abbas Khan
Email: abbaskhan@sjtu.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 1
Page No: 343-352
Abbas Khan
Email: abbaskhan@sjtu.edu.cn
Shoaib Saleem1, Abbas Khan2*, Mazhar Khan3, Abrar Mohammad Sayaf2, Syed Shujait Ali2
1National Center for Bioinformatics, Quaid-e-Azam University, Islamabad, Pakistan
2Center for Biotechnology and Microbiology, University of Swat, Pakistan
3The CAS Key Laboratory of Innate Immunity and Chronic Diseases, Hefei National Laboratory for Physical Sciences at Microscale, School of Life Sciences, CAS Center for Excellence in Molecular Cell Science, University of Science and Technology of China (USTC), Collaborative Innovation Center of Genetics and Development, Hefei, 230027, Anhui, China.
Shoaib Saleem: shoaibsaleem90@gmail.com; Abrar Mohammad Sayaf: chemdr07@gmail.com; Syed Shujait Ali: shujaitswati@uswat.edu.pk;
Zhijin Fan(fanzj@nankai.edu.cn)
Shoaib Saleem, Abbas Khan, Mazhar Khan, Abrar Mohammad Sayaf, Syed Shujait Ali, Reducing the pathological role of HIV Viruses by targeting the gateway GPR15: A Computational approach(2020)Journal of Computational Chemistry & Molecular Modeling 4(1) p:343-352
Here we reported recent advancement in the 3D structures of G protein-coupled receptors (GPCR) which act as major receptors in the spread of many diseases like HIV-1 and HIV-2. GPR15 is one among the GPCRs which has important role and act as important target for therapeutic agents. Consequently, we used comparatively/Homology modeling method to predict the 3D structure of GPR15 followed by the validation of the predicted structures by using Ramachandran Plot, Errat, Qmean and ProSA. An online server ProBiS was to predict the binding sites and ligands for them as showed similarities with other proteins binding site. Among the 22 ligands used for docking, UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine showed best binding energy than other compounds. This shows that UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine could be better for further bioassay. Using UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine for structure based virtual screening could help in the identification of further novel ligands with novel action, improved therapeutic efficacy and acceptable pharmacological properties.
Keywords: UK-432,097, 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine, G protein-coupled receptors (GPCR), Homology modeling, GPR15
G-Protein coupled receptors are present as embedded in membrane on the surface of the cell and are responsible for response to different stimulus like odorants, light, neurotransmitters, hormones, or pheromones can be used to activate these receptors (Dolphin, 1994; Flower, 1999; Uhlenbrock, Gassenhuber, & Kostenis, 2002). They carry out their function by interacting with heterotrimeric G proteins. These G-proteins coupled receptors are characterized by their 7 transmembrane domain structure. Over 800 G-proteins are divided into five families on the basis of its sequence analysis (Fredriksson, Lagerström, Lundin, & Schiöth, 2003). The malfunction of these receptors lead to many fatal diseases, making them important drug targets. GPCR remain an important target and is estimated that 45% of the drug target these receptors. It also constitute 25% of the top 100 selling drugs (I. H. G. S. Consortium, 2004; Drews, 2000; Flower, 1999; Takeda, Kadowaki, Haga, Takaesu, & Mitaku, 2002). Due to less or no availability of crystal and computational 3D structures of these receptors the use of structure-based approaches for drug design approach is limited (Archer, Maigret, Escrieut, Pradayrol, & Fourmy, 2003).
GPCR are also important to act as co-receptor and helps in the causing and spread of many diseases. GPR15 is one among the other receptors used by immunodeficiency viruses, HIV-1 and HIV-2 to cause HIV-AIDS in Humans and simian immunodeficiency virus from macaques (SIVmac) in Asian Macaques (Clavel et al., 1987; Gallo et al., 1984; Letvin et al., 1985). GPR15/BOB exhibits the primary structure of a 7-transmembrane domain protein and shares sequence identity with regions of the angiotensin II receptor and GPCRs CXCR2, CXCR4, CCR5, DEZ (chemerin receptor), GPR1 and APJ (apelin receptor)(Farzan et al., 1997; Heiber et al., 1996). The use of CD4 cells and chemokine receptors is a typical habit of Human immunodeficiency viruses (HIV) to enter the cell. CCR5 and CXCR4 were identified as co-receptors used by HIV virus for their entry to the cell (Deng et al., 1996; Dragic et al., 1996; Feng, Broder, Kennedy, & Berger, 1996). It has also been reported that GPR15 is used by many HIV2, SIVmac and some of HIV1 viruses to get entry to the cell, but the role of GPR15 is not clearly understood whether it expresses on the surface of virus infected cells or not (Blaak et al., 2005; Cilliers et al., 2005; Edinger et al., 1997; Krumbiegel & Kirchhoff, 1999; Mörner et al., 1999). GPR15/BOB is also responsible for HIV enteropathy condition characterized by malabsorption and increased intestinal permeability with diarrhea (Clayton et al., 2001). It is observed that TLR3 based infections increases the expression of GPR15 (Kiene et al., 2014). However, GPR15/BOB expression in chronic inflammatory disease has not been studied.
Due to the limited availability of the GPCR structures, the successful chemotherapy is restricted. Now, observing the importance and role of GPR15 more insightful study is required to understand and control the role of GPR15 in different diseases. Here, we reported the 3D homology model of GPR15 using computational tools. As there are very limited natural inhibitors available for GPCRs, so for GPR15 the inhibitors of other similar proteins having similar active site are docked. As early reported that 45% of the drug target these receptors so the use of such computational tools may help us to identify novel inhibitors that may possess pharmacological acceptable properties.
2.1. Software and hardware
To get a good quality model of GPR15, I-Tasser (Roy, Kucukural, & Zhang, 2010), Pyre 2 (Kelley, Mezulis, Yates, Wass, & Sternberg, 2015), Lomets (Wu & Zhang, 2007), Geno3D (Combet, Jambon, Deleage, & Geourjon, 2002), MOE (Vilar, Cozza, & Moro, 2008) and Modeler v 9.15(Webb & Sali, 2014) was used to generate a good homology model. The evaluation of the model was carried out using online servers including ERRAT (Colovos & Yeates, 1993), RAMPAGE (Lovell et al.), Qmean and ProSA(Wiederstein & Sippl, 2007). For docking MOE was used. Secondary structure prediction, active site prediction and other analysis of the receptor was carried out using online servers.
2.2. Templates selection and Sequence alignment
The complete amino acid sequence of GPR15 was retrieved from Uniprot (U. Consortium, 2014) using accession number P49685. The length of GPR 15 is 360 Amino acids. Using the primary sequence of GPR15 as query a PSI-BLAST was carried out to select suitable templates against the query. The PDB codes with the highest homology and coverage were retrieved from RCSB. Initially seven templates were used for manual model building. The alignment of the query sequence with the selected templates was carried out t-coffee online server (Di Tommaso et al., 2011).
2.3. Homology modeling
Homology modeling methodology was followed to construct the 3D homology model of GPR15. Homology modeling of GPR15 was carried out using different approaches including I-Tasser (Roy et al., 2010), Pyre 2 (Kelley et al., 2015), Lomets (Wu & Zhang, 2007), Geno3D (Combet et al., 2002), MOE (Vilar et al., 2008) and Modeler v 9.15(Webb & Sali, 2014). The final satisfactory model was generated using Modeler v 9.15, using the selected templates and also the models obtained from different servers. The loops were automatically refined using Modeler v 9.15. All the models were subjected to validation test for evaluation and finally a best model was selected for further analysis.
2.4. Secondary structure prediction and model validation
Self-Optimized Prediction Method (SOPMA)(Geourjon & Deleage, 1995) was used to predict the secondary structure of GPR15. The models obtained from I-Tasser, Pyre 2, Lomets, Geno3D, MOE and Modeler v 9.15were examined by using Ramachandran Plot, Errat, Qmean and ProSA. On the basis of good geometry the final model generated using Modeler v 9.15was further considered for analysis. The Ramachandran Plot revealed their Phi/Psi plot while ProSA relate the structure with X-ray and NMR structures.
2.5. Active site Prediction, Ligand Molecules and Molecular Docking
The active site of the finally refined model was predicted by using different online servers and finally the site finder tool of MOE to select the best site for docking. Structured based virtual screening protocol was followed to identify some inhibitors against GPR15 as no natural inhibitors are available for GPR15. Initially the inhibitors for other GPRs were used to identify some novel ligands. For the selection of ligand molecules an online server ProBiS (Clayton et al., 2001) was used which help to identify structurally similar binding site in different proteins and this ligands attached to those site. Using the results of ProBiS which identify some potential ligands on the basis of binding site similarities with other proteins. Before the docking all the identified ligands were subjected to protonation and energy minimization. A database for all the ligands were generated and stored as .mdb (molecular database). The protonation and minimization of the receptor was also carried out using MOE. Before the docking a database of these ligands was prepared using MOE. The parameters were set (Re-scoring function: London dG , placement: triangle matcher, Retain: 10, Refinement: Force field, and Re-scoring 2: London dG). Docking program of MOE provides correct conformation of the ligand so as to obtain minimum energy structure. After docking, S-score was considered the criteria to select best conformation for these ligands and these were then further studied to analyze the hydrogen bonding/π-π interactions through LigX tool of MOE.
3.1. Selection of Templates
After retrieving the primary sequence of GPR15 using accession no P49685 a PSI-BLAST was carried out to select good templates for the generation of 3D homology model of GPR15. Templates with highest homology and high sequence coverage were retrieved from RCSB and were utilized in the generation of final homology model. Initially seven templates were selected for generating the final model using Modeler v 9.15. Neuropeptide Receptor from Homo sapiens with 77% coverage and 25% homology, CXCR4 chemokine receptor with coverage 85% and homology 30% from Homo sapiens, N/OFQ Opioid Receptor from E.coli with 77% coverage and 30% of homology, CCR5 Chemokine Receptor from Clostridium pasteurianum with coverage of 82% and homology of 21%, human delta opioid 7 TM receptor from Homo sapiens with coverage of 80% and homology of 29%, human Angiotensin Receptor from homo sapiens with coverage of 83% and homology of 29% and Human Angiotensin Receptor from E.coli with coverage of 81% and homology of 32% were used to construct the final model of GPR15. Generally ≥30% of identity among query and templates are accepted for comparatively modeling (Forrest, Tang, & Honig, 2006). However, identity for membrane proteins ≥20% - 40% is widely accepted (Gao, 2009; Reddy, Vijayasarathy, Srinivas, Sastry, & Sastry, 2006). The requirement is showing that the templates we selected are far better than the required.
3.2. Sequence alignment & Secondary structure prediction
The alignment between the query sequence of GPR15 and the selected templates was carried out using an online server of t-coffee (Di Tommaso et al., 2011). The alignment of the query sequence with the selected templates are shown in the Figure 1. The prediction of secondary structure was carried out by using an online server SOPMA which resulted that 34.72% of the residues lies in Alpha helix, 27.50% in Extended strand (Ee), 8.06% in Beta Turn while 29.72% lies in random coil. This result shows that alpha helix occupied the largest portion of the protein, beta turn with the second on followed by extended strand. An online server TMpred (http://www.ch.embnet.org/cgi-bin/TMPRED_form_parser) was used to predict the number of transmembrane regions own by GPR15. It was showed that GPR15 contains 7 transmembrane regions which is showing that GPR15 is also possessing the same topology like other GPRs. This analysis suggested that GPR15 could be a good target for homology modeling. The depiction of the secondary structure regions are shown in the Figure 2 below.

Figure 1: Showing the alignment of the query sequence with the selected templates used in the generation of final 3D homology model of GPR15.
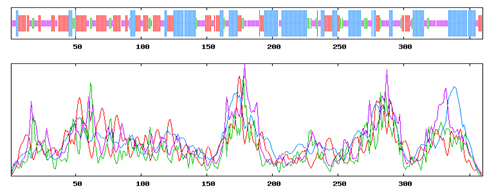
Figure 2: Showing the distribution of Alpha helix, beta turn, extended strand and random coil in the protein structure with different colours.
3.3. Model Building & Validation
Homology modeling methodology was followed to generate the best 3D model of GPR15. Initially many models were generated using online servers and evaluated but due to low quality they were just considered as templates for final model generation. Using I-Tasser (Roy et al., 2010), Phyre 2 (Kelley et al., 2015), Lomets (Wu & Zhang, 2007), Geno3D (Combet et al., 2002), MOE (Vilar et al., 2008) and Modeler v 9.15(Webb & Sali, 2014) to construct the models of GPR15. A model was also generated by using the selected templates through Modeler v 9.15v 9.15. The evaluation of the model was carried out using online servers including Errat (Colovos & Yeates, 1993), Rampage (Lovell et al.) and ProSA (Wiederstein & Sippl, 2007) but the results were not satisfactory. Finally the models obtained from different servers in combination with the selected templates were used to generate the best model of the protein. The final model generated after the use of multiple templates was subjected to model validation using different servers. The predicted model quality was evaluated by using ERRAT (Colovos & Yeates, 1993), RAMPAGE (Lovell et al.) ProSA(Wiederstein & Sippl, 2007) and QMEAN server (Benkert, Tosatto, & Schomburg, 2008). The final refined model of GPR15 is shown in the Figure 3.
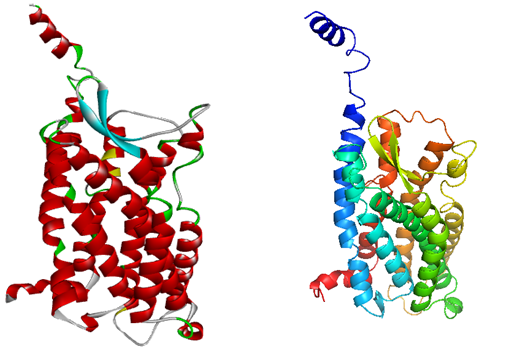
Figure 3. The figure is also showing the depiction of the final model generated by using Modeler v 9.15.
The stereo-chemical properties of the model was calculated by using an online server Rampage. On the evaluation of Ramachandran plot as shown in the Figure 4, 86.8% of the residues in were found favoured region, number of residues in allowed region were 9.4%, while the number of residues in outlier region 3.8%. By comparing these values with those of other built models using online servers, this model is the best model of all models generated. The Errat (Colovos & Yeates, 1993) analysis suggested that the overall quality factor was 95.83% and is considered the best model. The overall quality factor high score indicates the quality of the model. Hence, the quality of the final model is considered as the best as there are not as many residues over the quality indicator. Moreover, ProSA (Wiederstein & Sippl, 2007) analysis suggested that the model the native protein folding energy is good and was analyzed by utilizing Z-Score. The Z-score of the model was -4.26 which shows that the model built was within the range of standard requirements. Qmean server was further utilized to finally analyze the model quality. The Figure 5 is showing the quality of the final model generated. The ProSA server also showed that very few of the residues showed negative energy interaction while mostly lies in positive energy interaction.
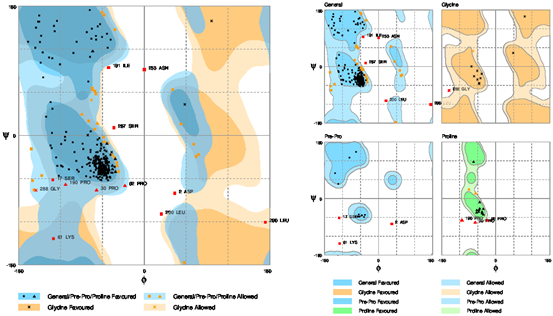
Figure 4: The given depiction of Ramachandran Plot showing the evaluation of the final model. The Plot showed (86.8%) 276 amino acids lies in the favored region, (9.4%) 30 amino acids plotted in allowed region while (3.8%) 12 amino acids lies in the outlier region.
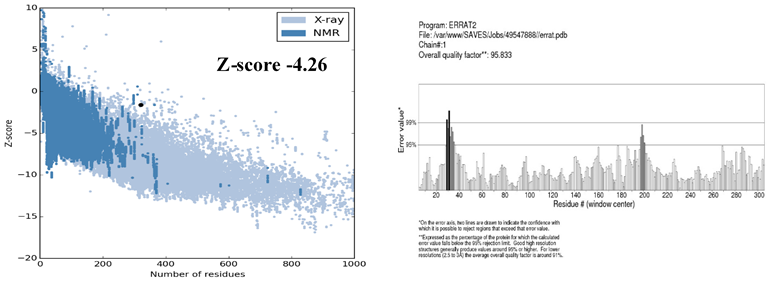
Figure 5: Showing the ProSA II and Errat value servers’ validation. The model Z-score was reported -4.26 and Errat overall quality factor was 95.8% which is reflecting the good quality of the model.
3.5. Molecular Docking
The ProBiS server identified morphinan, JDTic, 3-Quinuclidinyl benzilate, (E)-doxepin, tiotropium, UK-432,097, Stearic acid, CGS-21680, N-methyl scopolamine, Oleic acid, Cholesterol hemisuccinate, Retinal, Octyl β-D-glucopyranoside, Carazolol, Di-palmitoyl-3-sn-phosphatidylethanolamine, N-acetyl-d-glucosamine, 3,6,9,12-Tetraoxaicosan-1-ol, 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine, β-Carotene, Chlorophyll a, Tristearoylglycerol and Cobalamin as to have affinity towards the binding site of GPR15. These compounds were predicted on the basis of binding site similarities with other compounds. The final binding site used for docking was found by using Site finder tool of MOE as shown in the Figure 6. Around 30 conformations of these compounds showed different affinity which were analyzed by using binding energy, hydrogen bond forming residues and interacting residues. The statistical analysis indicated that among the 22 identified ligands only 1,2-dimyristoyl-sn-glycero-3-phosphocholine, 3,6,9,12-Tetraoxaicosan-1-ol, CGS-21680, Cholesterol hemisuccinate, Di-palmitoyl-3-sn-phosphatidylethanolamine, JDTic, N-acetyl-d-glucosamine, Oleic acid, Stearic Acid, Tristearoylglycerol and UK-432,097 were found to have good energy scoring. UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine as shown in the Figure 7 was found to have high score among all the docked ligands and thus showing a probability of influencing the activity of GPR15. The interaction of these ligands also confirm the residues of binding site as found common with other GPCRs. UK-432,097 total S-Score function is -14.7025 Kcal/mol and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine total S-Score function is -14.3749. This shows that UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine has good inhibitory activity on GPR15. Even though, some other compounds in this library are having better scoring but the UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine shows better interactions. So, we have demonstrated that, among the 22 identified ligands the binding capacity is highest for UK-432,097 and can be further used for further investigations.
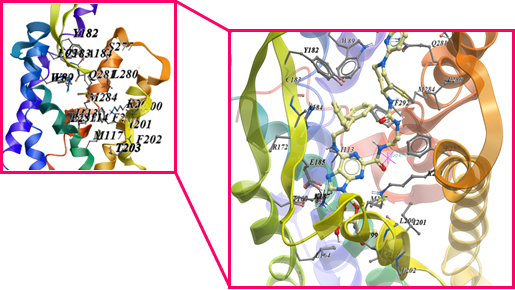
Figure 6: showing the predicted binding site and the docked ligand present at the binding site of GPR15.
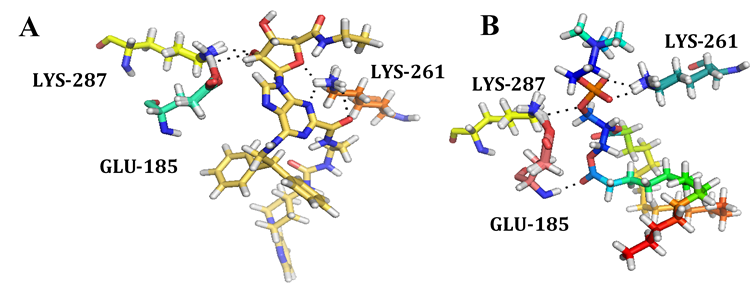
Figure 7: showing the interaction of (A) UK-432,097 and (B) 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine with GPR15 binding site residues.
Table: 1 showing the top most scoring compounds that were found to have good affinity with GPR15.
|
Conformation Number |
Compound Name |
Binding Energy |
Number of Hydrogen Bonds |
Interacting Residues |
|
2 |
1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine |
-14.3749 |
4 |
Cys 183, Tyr182,Glu 185, Trp 99, Lys 287, Lys 261 |
|
11 |
3,6,9,12-Tetraoxaicosan-1-ol |
-10.2027 |
4 |
Lys261, Phe 202 |
|
50 |
CGS-21680 |
-12.2044 |
4 |
Glu 93 , Lys 92 |
|
60 |
Cholesterol hemisuccinate |
-12.1302 |
2 |
Lys187 |
|
70 |
Di-palmitoyl-3-sn-phosphatidylethanolamine |
-12.4809 |
2 |
His28 |
|
74 |
JDTic |
-11.3645 |
2 |
Glu185, Trp85 |
|
90 |
N-acetyl-d-glucosamine |
-9.06826 |
3 |
Glu185, Ser114, Lys261 |
|
100 |
Oleic acid |
-9.58019 |
2 |
Glu285 |
|
130 |
Tristearoylglycerol |
-13.2613 |
2 |
Glu185 |
|
140 |
UK-432,097 |
-14.7025 |
5 |
Glu 185, Tyr 182, Lys 287, Lys 261 |
In this work, we have constructed a 3D model of G-protein coupled receptor 15 of Homo sapiens, which plays an important role in HIV-1 and HIV-2 infection. A refined model was obtained after energy minimization. The validation of protein structure through Ramachandran Plot, Errat, Qmean and ProSA validates that the protein is stable enough. The stable structure is further used for docking with the identified ligands using the binding site similarity. Docking results indicated that there exist a good affinity between the ligands and GPR15. The interaction between the domain and the inhibitors proposed in this study are useful for understanding the potential mechanism of domain and the inhibitor binding. The hydrogen bonds play important role for the structure and function of biological molecule in this study, and we found that CYS 183, TYR182, TRP 99, LYS 261, PHE 202, HIS 28 and LYS 187 of GPR15 are important for strong hydrogen bonding interaction with these inhibitors. Among the 22 ligands used for docking, UK-432,097 and 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine showed best binding energy than other compounds. The total S-score function is high for UK-432,097 when compared to other ligands including 1,2-Dimyristoyl-Sn-Glycero-3-Phosphocholine. According to our investigations from the docking results UK-432,097 can act as better GPR15 antagonist. Future investigation of other theoretical studies and experimental studies may confirm that UK-432,097 as a potent inhibitor.
Archer, E., Maigret, B., Escrieut, C., Pradayrol, L., & Fourmy, D. (2003). Rhodopsin crystal: new template yielding realistic models of G-protein-coupled receptors? Trends in pharmacological sciences, 24(1), 36-40. 00009-3
View ArticleBenkert, P., Tosatto, S. C., & Schomburg, D. (2008). QMEAN: A comprehensive scoring function for model quality assessment. Proteins: Structure, Function, and Bioinformatics, 71(1), 261-277. PMid:17932912
View Article PubMed/NCBIBlaak, H., Boers, P., Gruters, R., Schuitemaker, H., Van Der Ende, M., & Osterhaus, A. (2005). CCR5, GPR15, and CXCR6 are major coreceptors of human immunodeficiency virus type 2 variants isolated from individuals with and without plasma viremia. Journal of virology, 79(3), 1686-1700. PMid:15650194
View Article PubMed/NCBICilliers, T., Willey, S., Sullivan, W. M., Patience, T., Pugach, P., Coetzer, M., . . . Clapham, P. (2005). Use of alternate coreceptors on primary cells by two HIV-1 isolates. Virology, 339(1), 136-144. PMid:15992849
View Article PubMed/NCBIClavel, F., Mansinho, K., Chamaret, S., Guetard, D., Favier, V., Nina, J., . . . Montagnier, L. (1987). Human immunodeficiency virus type 2 infection associated with AIDS in West Africa. New England Journal of Medicine, 316(19), 1180-1185. PMid:3472076
View Article PubMed/NCBIClayton, F., Kotler, D. P., Kuwada, S. K., Morgan, T., Stepan, C., Kuang, J., . . . Fantini, J. (2001). Gp120-induced Bob/GPR15 activation: a possible cause of human immunodeficiency virus enteropathy. The American journal of pathology, 159(5), 1933-1939. 63040-4
View ArticleColovos, C., & Yeates, T. O. (1993). Verification of protein structures: patterns of nonbonded atomic interactions. Protein Science, 2(9), 1511-1519. PMid:8401235
View Article PubMed/NCBICombet, C., Jambon, M., Deleage, G., & Geourjon, C. (2002). Geno3D: automatic comparative molecular modelling of protein. Bioinformatics, 18(1), 213-214. PMid:11836238
View Article PubMed/NCBIConsortium, I. H. G. S. (2004). Finishing the euchromatic sequence of the human genome. Nature, 431(7011), 931-945. PMid:15496913
View Article PubMed/NCBIConsortium, U. (2014). UniProt: a hub for protein information. Nucleic acids research, gku989.
Deng, H., Liu, R., Ellmeier, W., Choe, S., Unutmaz, D., Burkhart, M., . . . Hill, C. M. (1996). Identification of a major co-receptor for primary isolates of HIV-1. PMid:8649511
View Article PubMed/NCBIDi Tommaso, P., Moretti, S., Xenarios, I., Orobitg, M., Montanyola, A., Chang, J.-M., . . . Notredame, C. (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic acids research, 39(suppl 2), W13-W17. PMid:21558174
View Article PubMed/NCBIDolphin, A. (1994). The G protein-linked receptor facts book. Neuroscience, 62(4), 1309. 90364-6
View ArticleDragic, T., Litwin, V., Allaway, G. P., Martin, S. R., Huang, Y., Nagashima, K. A., . . . Moore, J. P. (1996). HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. PMid:8649512
View Article PubMed/NCBIDrews, J. (2000). Drug discovery: a historical perspective. Science, 287(5460), 1960-1964. PMid:10720314
View Article PubMed/NCBIEdinger, A. L., Amedee, A., Miller, K., Doranz, B. J., Endres, M., Sharron, M., . . . Murphey-Corb, M. (1997). Differential utilization of CCR5 by macrophage and T cell tropic simian immunodeficiency virus strains. Proceedings of the National Academy of Sciences, 94(8), 4005-4010. PMid:9108095
View Article PubMed/NCBIFarzan, M., Choe, H., Martin, K., Marcon, L., Hofmann, W., Karlsson, G., . . . Sullivan, N. (1997). Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. The Journal of experimental medicine, 186(3), 405-411. PMid:9236192
View Article PubMed/NCBIFeng, Y., Broder, C. C., Kennedy, P. E., & Berger, E. A. (1996). HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science, 272(5263), 872-877. PMid:8629022
View Article PubMed/NCBIFlower, D. R. (1999). Modelling G-protein-coupled receptors for drug design. Biochimica et Biophysica Acta (BBA)-Reviews on Biomembranes, 1422(3), 207-234. 00006-4
View ArticleForrest, L. R., Tang, C. L., & Honig, B. (2006). On the accuracy of homology modeling and sequence alignment methods applied to membrane proteins. Biophysical journal, 91(2), 508-517. PMid:16648166
View Article PubMed/NCBIFredriksson, R., Lagerström, M. C., Lundin, L.-G., & Schiöth, H. B. (2003). The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular pharmacology, 63(6), 1256-1272. PMid:12761335
View Article PubMed/NCBIGallo, R. C., Salahuddin, S. Z., Popovic, M., Shearer, G. M., Kaplan, M., Haynes, B. F., . . . Safai, B. (1984). Frequent detection and isolation of cytopathic retroviruses (HTLV-III) from patients with AIDS and at risk for AIDS. Science, 224(4648), 500-503. PMid:6200936
View Article PubMed/NCBIGao, C. (2009). Computational studies on membrane protein structures and protein-ligand binding affinities. University of Rochester.
Geourjon, C., & Deleage, G. (1995). SOPMA: significant improvements in protein secondary structure prediction by consensus prediction from multiple alignments. Computer applications in the biosciences: CABIOS, 11(6), 681-684. PMid:8808585
View Article PubMed/NCBIHeiber, M., Marchese, A., Nguyen, T., Heng, H. H., George, S. R., & O'Dowd, B. F. (1996). A novel human gene encoding a G-protein-coupled receptor (GPR15) is located on chromosome 3. Genomics, 32(3), 462-465. PMid:8838812
View Article PubMed/NCBIKelley, L. A., Mezulis, S., Yates, C. M., Wass, M. N., & Sternberg, M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. [Protocol]. Nat. Protocols, 10(6), 845-858. doi: 10.1038/nprot.2015.053 PMid:25950237
View Article PubMed/NCBIKiene, M., Rethi, B., Jansson, M., Dillon, S., Lee, E., Lantto, R., . . . Chiodi, F. (2014). Toll-like receptor 3 signalling up-regulates expression of the HIV co-receptor G-protein coupled receptor 15 on human CD4+ T cells. PloS one, 9(2), e88195. PMid:24558379
View Article PubMed/NCBIKrumbiegel, M., & Kirchhoff, F. (1999). Coreceptor usage of BOB/GPR15 and Bonzo/STRL33 by primary isolates of human immunodeficiency virus type 1. Journal of general virology, 80(5), 1241-1251. PMid:10355771
View Article PubMed/NCBILetvin, N., Daniel, M., Sehgal, P., Desrosiers, R., Hunt, R., Waldron, L., . . . King, N. (1985). Induction of AIDS-like disease in macaque monkeys with T-cell tropic retrovirus STLV-III. Science, 230(4721), 71-73. PMid:2412295
View Article PubMed/NCBILovell, S., Davis, I., Adrendall, W., de Bakker, P., Word, J., & Prisant, M. Richardson, JS, Richardson, DC 2003. Structure validation by C alpha geometry: 18 phi, psi and C beta deviation. Proteins, 50, 437-450. PMid:12557186
View Article PubMed/NCBIMörner, A., Björndal, Å., Albert, J., KewalRamani, V. N., Littman, D. R., Inoue, R., . . . Björling, E. (1999). Primary human immunodeficiency virus type 2 (HIV-2) isolates, like HIV-1 isolates, frequently use CCR5 but show promiscuity in coreceptor usage. Journal of virology, 73(3), 2343-2349. PMid:9971817
View Article PubMed/NCBIReddy, C. S., Vijayasarathy, K., Srinivas, E., Sastry, G. M., & Sastry, G. N. (2006). Homology modeling of membrane proteins: a critical assessment. Computational biology and chemistry, 30(2), 120-126. PMid:16540373
View Article PubMed/NCBIRoy, A., Kucukural, A., & Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. [10.1038/nprot.2010.5]. Nat. Protocols, 5(4), 725-738. PMid:20360767
View Article PubMed/NCBITakeda, S., Kadowaki, S., Haga, T., Takaesu, H., & Mitaku, S. (2002). Identification of G protein-coupled receptor genes from the human genome sequence. FEBS letters, 520(1), 97-101. 02775-8
View ArticleUhlenbrock, K., Gassenhuber, H., & Kostenis, E. (2002). Sphingosine 1-phosphate is a ligand of the human gpr3, gpr6 and gpr12 family of constitutively active G protein-coupled receptors. Cellular signalling, 14(11), 941-953. 00041-4
View ArticleVilar, S., Cozza, G., & Moro, S. (2008). Medicinal chemistry and the molecular operating environment (MOE): application of QSAR and molecular docking to drug discovery. Current topics in medicinal chemistry, 8(18), 1555-1572. PMid:19075767
View Article PubMed/NCBIWebb, B., & Sali, A. (2014). Comparative protein structure modeling using Modeller. Current protocols in bioinformatics, 5.6. 1-5.6. 32. PMid:25199792
View Article PubMed/NCBIWiederstein, M., & Sippl, M. J. (2007). ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic acids research, 35(suppl 2), W407-W410. PMid:17517781
View Article PubMed/NCBIWu, S., & Zhang, Y. (2007). LOMETS: a local meta-threading-server for protein structure prediction. Nucleic acids research, 35(10), 3375-3382. PMid:17478507
View Article PubMed/NCBI