COMPARISON OF THE QUALITY OF DNA EXTRACTED FROM BUCCAL SWABS USING SINGLE STEP AND MULTI-STEP PROCEDURES
Corresponding Author
EMAIL: rsantos@southuniversity.edu
Affiliation
Jomarie Ortiz-Rivera and Roseane Maria Maia Santos
Department of Pharmaceutical Sciences, South University School of Pharmacy, Savannah, GA, USA
Article Reviewed By:
Yamunah Devi Apalasamy(yamuna219@yahoo.com)
Citation
Roseane Maria Maia Santos, COMPARISON OF THE QUALITY OF DNA EXTRACTED FROM BUCCAL SWABS USING SINGLE STEP AND MULTI-STEP PROCEDURES(2017)SDRP Journal of Earth Sciences & Environmental Studies 2(4)
Abstract
Genomic DNA has become a very important source for multiple purposes in research projects. Therefore, the analysis of the quality of the extraction of DNA using different specimen collection methods; are part of every materials and method section in current papers. The objective of this study was to evaluate which protocol extracts the highest amount with better quality of DNA from human buccal swabs. There are two different ways to harvest DNA from buccal swabs: single or multi-step procedure. Our study compared the quality of the DNA extracted using these two different methods of DNA extraction; though the determination spectroscopic measurements at 280, 260, and 230 nm wavelengths and the suitability for application in genotyping assays.
Introduction
Genomic DNA has become a very important source for multiple purposes in research projects. Therefore, the analysis of the quality of the extraction of DNA using different specimen collection methods; are part of every materials and method section in current papers (1-7). The objective of this study was to evaluate which protocol extracts the highest yield with better quality of DNA from human buccal swabs. There are two different ways to harvest DNA from buccal swabs: single or multi-step procedure. Our study compared the quality of the DNA extracted using these two different methods of DNA extraction; though the determination spectroscopic measurements at 280, 260, and 230 nm wavelengths and the suitability for application in genotyping assays.
Materials & Methods
Buccal cotton swabs (Isohelix Part # SK-2S, Boca Scientific, Boca Raton, FL, USA) were collected from 7 healthy volunteers. Each participant was given two collection kits containing the DNA buccal swabs, and an instruction video detailing the manufacturer’s directions of how to collect the sample. The manufacturer recommends collecting the buccal sample during a period of 60 seconds. Our study compared the quality of the DNA harvest from 20“and 60” collection time and extracted by simple and multi-step procedures. Two commercial DNA extraction methods were tested. Method A (Isohelix, part # BEK-50, Boca Scientific, Boca Raton, FL, USA) uses a single-step procedure, and Method B (Isohelix, part # BEK-50, Boca Scientific, Boca Raton, FL, USA) (Isohelix, part # BPK-50, Boca Scientific, Boca Raton, FL, USA) using a multi-step procedure.
DNA Extraction Method
Method A: Single-Step
Buccal cotton swabs were released from the stick into a clean 2 mL microcentrifuge tube and 400 µL of Buccalyse solution (BEK) was added to each sample to extract the proteins from the swabs and vortexed briefly. A 100 µL of the lysed solution of each sample was transferred into a clean 8-tubes PCR strip. The tubes were capped and we utilize a thermocycler programmed with the following steps: 70°C for 15 minutes, 95°C for 2 minutes, 20°C for 2 minutes. The samples were stored at 4°C to be analyzed 48 hours later. At this point the DNA samples were ready to be use in downstream applications (8, 9).
Method B: Multi-Step
Buccal cotton swabs were released from the stick into a clean 2 mL centrifuge tube and 500 µL of buccal lysis and stabilization buffer (BLS) was added to each sample to stabilize the DNA. The tubes were sealed, and vortexed briefly, and stored at 4°C. Next, 20 µL of Proteinase K (PK) were added to all samples, vortexed briefly and incubated in a pre-heated 60°C heat block for 30 minutes. Then approximately 400 µL of the solution was transferred into a 1.5 mL microcentrifuge tube and 400 µL of the precipitation buffer (BP) were added, briefly vortexed, and centrifuged at maximum speed (13.2K rpm/12,000 x g) for 10 minutes. At this point we could see a white pellet of genetic material. The supernatant was removed by pipetting and reversing the tube carefully into a paper towel. Fifty microliters of the re-hydration buffer (TE) was added to each tube to obtain a concentrated DNA solution. The samples were vortexed for 30 seconds to fully resuspend the white pellet and left at room temperature for the DNA to re-hydrate. The final step consisted of sample centrifugation for 15 minutes at maximum speed (13.2K rpm/12,000 x g) to remove any undissolved impurity. At this point the DNA samples were ready to be use in downstream applications (8, 9).
Nucleic acid quantification
Quantification and evaluation of the quality of the gDNA extracted through the two different methods and the two sample time collections were done using the NanoDrop® Spectrophotometer 2000c (Thermo Scientific, Wilmington, DE).
Real-Time PCR genotyping method
The genotype of the CYP1A2 enzyme (rs762551) was assayed using TaqMan® Drug Metabolism Assay (Applied Biosystems, Carlsbad, CA). The TaqMan® genotype technology uses two sequence specific primers to amplify the DNA fragment of interest and two allelic-specific TaqMan® Minor Grove Binder (MGB) probes to detect the two polymorphic alleles. Allele-1 and Allele-2 are discriminated by the presence of the fluorescence of VIC® dye and FAM® dye respectively. The fluorescence emission for the two reporter dyes is normalized in reference to a passive dye (ROX® dye). The real-Time PCR genotype assay was run on StepOnePlus™ (Applied Biosystems, Carlsbad, CA).
Results
The concentration of DNA obtained with single-step method ranged from 105.3 - 302.6 ng/ μL for a 60” collection time (σ= 66.2); and 158.6 - 299.9 ng/ μL for 20” collection time (σ= 42.1); averaging 178.6 and 207.28 ng/ μL respectively. The concentration of DNA obtained with multi-step method ranged from 30.9 - 84.45 ng/ μL (381.6 ng/μL, outlier) for a 60“collection time (σ= 19.6); and 44.0 - 114.6 ng/ μL for 20” collection time (σ= 22.8); averaging 53.55 and 66.92 ng/μL respectively (Table 1). The A260/280 ratio is generally used to determine protein contamination of a nucleic acid sample. The aromatic proteins have a strong UV absorbance at 280 nm. For pure RNA and DNA, A260/280 ratio should be around 2.0 - 1.8, respectively. A lower ratio indicates the sample is protein contaminated. The presence of protein contamination may influence downstream applications that use the nucleic acid samples.
The A260/280 ratio obtained with the single-step method ranged from 0.9 - 1.1 (σ= 0.1); and the A260/230 ratio ranged from 0.1 - 0.3 for 60“collection time (σ= 0.2). The A260/280 ratio obtained with the single-step method ranged from 1.0 - 1.1 (σ= 0.1); and the A260/230 ratio ranged from 0.2 - 0.4 for 20“collection time (σ= 0.1). The A260/280 ratio obtained with the multi-step method ranged from 1.4 - 1.8 (σ= 0.1); and the A260/230 ratio ranged from 0.4 - 0.8 for 60 “collection time (σ= 0.2). The A260/280 ratio obtained with the multi-step method ranged from 1.4 - 1.9 (σ= 0.2); and the A260/230 ratio ranged from 0.2 - 0.9 for 20 “collection time (σ= 0.2) (Table 1).
Discussion
This study resulted from a preliminary comparison between different kits to obtain DNA for genotyping purposes. We observed that there were a significant DNA yield and quality difference, when we utilized a single step procedure compared with a multi-step. In order to decide which procedure to adopt, we enrolled seven healthy volunteers that agreed to donate buccal swab samples to perform the study. Also, the duration of time to collect the buccal swabs was tested for 20 and 60 seconds, targeting to evaluate the possibility to expedite the collection process without loss of DNA quality. Therefore, the strength of this study was to bring to discussion the methods of collection of buccal swabs as well as the techniques to harvest the DNA from those samples; without compromising the results. The limitation of the study is the sample size and lack of comparison with other kits available in the market. The popularity of self-collection of buccal swabs with an increasing number of purposes, such as ancestry and search for DNA mutations, this study might prove to be useful and bring to attention of researchers about the collection procedure and DNA quality and quantity obtained by current DNA extraction methods.
Conclusion
The modification of the collection time from 60 seconds (according to manufacturer’s instructions) to 20 seconds prove to be reliable since using single-step method the DNA yield average 208.3 ng/ μL and 207.3 ng/ μL respectively. These results were reproduced when using multi-step procedure, averaging 64.3 ng/μL and 66.9 ng/μL for 60 and 20 seconds collection time respectively.
The modification of the collection time from 60 seconds (according to manufacturer’s instructions) to 20 seconds prove to be reproducible in terms of DNA quality since the ratio A260/280 with single-step method showed no significant variation in the standard deviation (Table1). The ratio A260/230 relates the amount of protein impurities (230 nm absorbance) with organic compounds that have strong absorbance around 225 nm (mostly due to formula components of the extractant reagents). Therefore, the ratios obtained for A260/230 ratio in our experiments, which ranged between 0.1 - 0.9 can be explained by the higher content of the residual reagents than actually protein impurities.
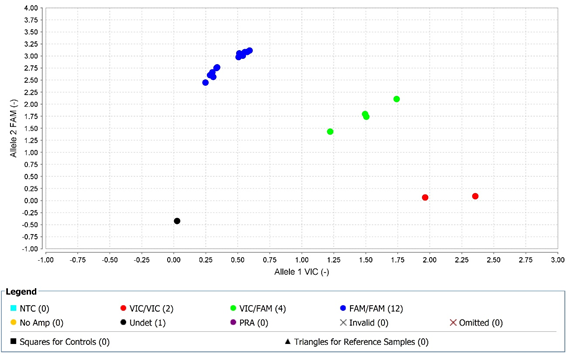
We selected genotyping application as a method to evaluate the quality of the DNA obtained from both procedures with different collection times. Figure 1 shows that TaqMan® Drug Metabolism Assay for CYP1A2 rs762551 discriminated both alleles clustering the three different genotypes on all samples (homozygous allele 1; homozygous allele 2 and heterozygous). No invalid or undetermined genotype was found.
Our conclusion is that single-step method presents higher DNA yield with lower quality and the multi-step method produce lower DNA yield with higher quality. However, both procedures can be utilized to obtain DNA from buccal cotton swabs and successfully perform genotyping assays. In addition, the time of collection can be reduced to a third without compromising the results.
Images and Tables
|
Sample ID |
DNA concentration after collection (ng/µL) |
280/260 ratio |
260/230 ratio |
Collection Time (seconds) |
Method |
Allelic Discrimination CYP 1A2 rs 762551 |
|
1 |
220.05 |
1.015 |
0.265 |
60 |
Single-Step |
Heterozygous • |
|
3 |
105.3 |
0.945 |
0.145 |
60 |
Single-Step |
Homozygous Allele 2 • |
|
4 |
162.6 |
1.00 |
0.2 |
60 |
Single-Step |
Homozygous Allele 2 • |
|
5 |
186.75 |
1.02 |
0.23 |
60 |
Single-Step |
Homozygous Allele 2 • |
|
6 |
272.62 |
1.045 |
0.34 |
60 |
Single-Step |
Homozygous Allele 2 • |
|
7 |
302.55 |
1.145 |
0.17 |
60 |
Single-Step |
Homozygous Allele 1 • |
|
1 |
381.6 |
1.58 |
0.34 |
60 |
Multi-Step |
Heterozygous • |
|
2 |
30.9 |
1.75 |
0.53 |
60 |
Multi-Step |
Homozygous Allele 2 • |
|
3 |
82.7 |
1.44 |
0.36 |
60 |
Multi-Step |
Homozygous Allele 2 • |
|
5 |
84.45 |
1.735 |
0.66 |
60 |
Multi-Step |
Homozygous Allele 2 • |
|
6 |
56.35 |
1.575 |
0.42 |
60 |
Multi-Step |
Homozygous Allele 2 • |
|
7 |
66.95 |
1.725 |
0.775 |
60 |
Multi-Step |
Homozygous Allele 1 • |
|
1 |
197.9 |
1.08 |
0.24 |
20 |
Single-Step |
Heterozygous • |
|
2 |
158.6 |
1.03 |
0.20 |
20 |
Single-Step |
Homozygous Allele 2 • |
|
3 |
211.2 |
1.02 |
0.26 |
20 |
Single-Step |
Homozygous Allele 2 • |
|
4 |
216.0 |
0.97 |
0.28 |
20 |
Single-Step |
Homozygous Allele 2 • |
|
5 |
192.3 |
1.13 |
0.24 |
20 |
Single-Step |
Homozygous Allele 2 • |
|
6 |
175.1 |
1.07 |
0.22 |
20 |
Single-Step |
Homozygous Allele 2 • |
|
7 |
299.9 |
1.10 |
0.37 |
20 |
Single-Step |
Homozygous Allele 1 • |
|
1 |
114.6 |
1.89 |
0.92 |
20 |
Multi-Step |
Heterozygous • |
|
2 |
59.3 |
1.43 |
0.29 |
20 |
Multi-Step |
Homozygous Allele 2 • |
|
3 |
44.0 |
1.46 |
0.26 |
20 |
Multi-Step |
Homozygous Allele 2 • |
|
4 |
44.1 |
1.42 |
0.23 |
20 |
Multi-Step |
Homozygous Allele 2 • |
|
5 |
81.9 |
1.51 |
0.41 |
20 |
Multi-Step |
Homozygous Allele 2 • |
|
6 |
61.2 |
1.52 |
0.39 |
20 |
Multi-Step |
Homozygous Allele 2 • |
|
7 |
63.4 |
1.45 |
0.34 |
20 |
Multi-Step |
Homozygous Allele 1 • |
Allele 1= mutant type recessive; allele 2= wild type dominant;
References
Burger MF, Song EY, Schumm JW. Buccal DNA samples for DNA typing: new collection and processing methods. Biotechniques 2005;39(2):257-61. PMid:16116799
View Article PubMed/NCBIHerraez DL, Stoneking M. High fractions of exogenous DNA in human buccal samples reduce the quality of large-scale genotyping. Anal Biochem 2008;383(2):329-31. PMid:18804445
View Article PubMed/NCBIKing IB, Satia-Abouta J, Thornquist MD, Bigler J, Patterson RE, Kristal AR, et al. Buccal cell DNA yield, quality, and collection costs: comparison of methods for large-scale studies. Cancer Epidemiol Biomarkers Prev 2002;11(10 Pt 1):1130-3. PMid:12376522
PubMed/NCBILivy A, Lye S, Jagdish CK, Hanis N, Sharmila V, Ler LW, et al. Evaluation of quality of DNA extracted from buccal swabs for microarray based genotyping. Indian J Clin Biochem 2012;27(1):28-33. PMid:23277709
View Article PubMed/NCBIRichards B, Skoletsky J, Shuber AP, Balfour R, Stern RC, Dorkin HL, et al. Multiplex PCR amplification from the CFTR gene using DNA prepared from buccal brushes/swabs. Hum Mol Genet 1993;2(2):159-63. PMid:7684637
View Article PubMed/NCBIWalker AH, Najarian D, White DL, Jaffe JF, Kanetsky PA, Rebbeck TR. Collection of genomic DNA by buccal swabs for polymerase chain reaction-based biomarker assays. Environ Health Perspect 1999;107(7):517-20. PMid:10378997
View Article PubMed/NCBIZheng S, Ma X, Buffler PA, Smith MT, Wiencke JK. Whole genome amplification increases the efficiency and validity of buccal cell genotyping in pediatric populations. Cancer Epidemiol Biomarkers Prev 2001;10(6):697-700. PMid:11401921
PubMed/NCBIMcMichael GL, Gibson CS, O'Callaghan ME, Goldwater PN, Dekker GA, Haan EA, et al. DNA from buccal swabs suitable for high-throughput SNP multiplex analysis. J Biomol Tech 2009;20(5):232-5. PMid:19949693
PubMed/NCBIWong LS, Huzen J, de Boer RA, van Gilst WH, van Veldhuisen DJ, van der Harst P. Telomere length of circulating leukocyte subpopulations and buccal cells in patients with ischemic heart failure and their offspring. PLoS One 2011;6(8):e23118. PMid:21876736 PMCid:PMC3158078
View Article PubMed/NCBI
