Jin-Ying Gou
Tel: 86-21-31246758
Email: jygou@fudan.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
Jin-Ying Gou
Tel: 86-21-31246758
Email: jygou@fudan.edu.cn
Meng-An Kuang1, Peng-Cheng Zhou1, Awais Rasheed2, and Jin-Ying Gou1,*
1 School of Life Sciences, Fudan University, Shanghai 200438, China
2 Plant Sciences, Quaid-i-Azam University, Islamabad 45320, Pakistan
Natalia(almasia.natalia@inta.gob.ar)
Zhou S(zhoushuobio@163.com)
Ni Z(nizhiyong@126.com)
Yu Y(yuyuehua1213@sina.com)
Meng-An Kuang, Peng-Cheng Zhou, Jin-Ying Gou, Genome-wide analyses highlight wheat Cin-namoyl-CoA reductases' potential against abiotic and biotic stresses (2021) Journal of Plant Science 5(1) :245-260
Lignin is a crucial component of the cell wall, the first line of plant defense against abiotic and biotic stresses. Cinnamoyl-CoA reductase (CCR) catalyzes the first committed step for lignin monolignol synthesis. Potential roles of CCRs in stress responses are still elusive in bread wheat. Here, 114 wheat CCRs were identified from wheat genome information and were categorized into seven groups. We investigated the phylogenetic relationship, conserved protein motifs, chromosome localization, and gene structure to gain insight and show the homologous relationships among these CCRs. Using open transcriptome data, we identified 11 ubiquitous, and 103 tissue specific expressed CCRs genes in wheat. Gene expression increase in groups 2, 6, and 7 at the early infection stage and group 1 at the late Puccinia striiformis f. sp. tritici infection stage. Fusarium graminearum infection significantly induced the group 3 CCR genes. Finally, three CCR genes significantly increased at the late stage of drought treatment with significantly more lignin contents. This study provided genome-wide identification and overall transcriptome insights of wheat CCRs, highlighting their potential role in wheat stress resistance.
Keywords: Wheat; lignin; Cinnamoyl-CoA reductase; biotic stress; abiotic stress; drought
Wheat (Triticum aestivum L.) roughly provides one-fifth of the proteins and calories for human beings [1]. Now, over 700 million tons of wheat are produced annually worldwide (http://www.fao.org/faostat/en/). However, the yield and quality of wheat are under severe threat from extreme environmental stresses, both biotic and abiotic.
The abiotic stresses, including cold, heat, drought, and salinity, all significantly affect the yield of wheat and weaken global food security [2-4]. The total chlorophyll concentration decreased under low temperature, and the maximum quantum yield of the PSII (Fv/Fm) was significantly lower than control plants [5]. At high temperatures, the decrease of detoxifying enzymes leads to the accumulation of reactive oxygen species, which accompanied the thylakoid membrane lipid composition and cell organelle damages in wheat leaf cells to reduce the photosynthetic rate [6]. The drought stress reduced water potential and chlorophyll contents in flag leaves, decreased the number of cells in the endosperm, reduced the grain-filling rate and grain weight of wheat [7].
Wheat is widely grown in the world with different environments and faces the threat of a wide range of pests and diseases, including foliar and stem diseases, seed transmitted diseases, and soil-borne diseases [8]. Between 2000 and 2012, stripe rust expanded in 35 (55%) of the 64 countries, which constituted 76.8% of the world’s wheat crop in 2012 [9]. About 5.47 million tonnes of wheat were estimated to be lost to the pathogen each year, equivalent to a loss of US$979 million per year [10]. Fusarium graminearum (sexual stage: Gibberella zeae) causes the devastating head blight or scab disease on wheat and causes significant crop and quality losses with the trichothecene mycotoxins (e.g., deoxynivalenol), which constitutes a significant threat to human and animal health [11]. Change of lignin content and provide the first line of defense in plants against the pathogenic stress [12].
Lignin, one of the major compositions in the cell wall, is a phenolic polymer derived mainly from hydroxycinnamyl alcohols and is ubiquitously present in plants [13]. Lignin plays a critical role in the morphological formation of plants and works as the structural basis for structural rigidity for tracheophytes to stand upright and strengthens water-conducting tracheary elements to withstand the negative pressure generated during transpiration [14]. Lignin reinforces the cell walls, facilitates water transport, and acts as a physical barrier to pathogens [15]. Both lignin content and syringyl lignin correlated to tobacco (Nicotiana tabacum L.)’s resistance against Phytophthora parasitica var. nicotianae and Pseudomonas solanacearum [16]. 4-coumaroyl CoA is the central metabolite of the phenylpropanoids pathway and works as either the direct precursor for flavonoid or para-hydroxy-phenyl lignin biosynthesis or is fed into the production of increasingly methoxylated guaiacyl (G)- and syringyl (S)-monolignols [17].
Cinnamoyl-CoA reductase (CCR) catalyzes the first committed step for lignin monolignol synthesis [18]. Together with cinnamyl alcohol dehydrogenase (CAD), CCR converts p-coumaroyl-CoA, feruloyl-CoA, and sinapoyl-CoA into p-coumaryl, coniferyl, and sinapyl alcohols, the monolignol precursors for H, G, and S lignin [19]. Based on the expression profiles, CCR contains two subfamilies: one devoted to developmental lignification and the other involved in synthesizing defense-related compounds [20]. In rice (Oryza sativa L.), a sphingolipid elicitor induced OsCCR1, whose encoded protein was bound and activated by OsRac1, one of the Rac/Rop family small GTPases to accumulated lignin and ROS [21]. The expressions of OsCCR17 and 21 were induced in response to biotic and abiotic stresses, such as Magnaporthe grisea and Xanthomonas oryzae pv. oryzae (Xoo) infections, UV-irradiation, and high salinity, suggesting a role of these genes in rice defense-related processes [22]. Under Polyethylene glycol (PEG) stress, CCR1 increased in foxtail millet (Setaria italica L.) during phase I but decreased in phase II and III under PEG stress. Simultaneously, a higher amount of cinnamic acid accumulated in germinating seeds under PEG than compared to control treatment, suggesting phenylpropanoid’s involvement in this process [23]. In the developing seedlings of Leucaena leucocephala, an increase in lignification was observed in mannitol-treated stems and the corresponding CCR protein accumulated at a level higher than control [24]. Compared with the extensive studies of CCR’s biological functions in response to biotic and abiotic stresses in model plants such as Arabidopsis and rice, little progress was made to elucidate the biological function of CCR genes in wheat.
Here we used the current genomic information and analyzed the CCR gene family in wheat. We investigated the location, gene structure, cis-elements in the promoters, and CCR family genes’ expression during wheat development and response to different stresses. Several CCR genes showed a strong induction upon pathogen infection, indicating their potential involvement in disease resistance. Moreover, we identified several CCR genes responsible for tolerance to osmotic stress simulated by PEG treatment. These results provided useful information for the functional study of the CCR gene in wheat against environmental stresses.
2.1. Genome-wide identification of CCR family genes in wheat
The genome data of wheat was downloaded from the Ensemble Plants database (http://plants.ensembl.org/Triticum_aestivum/Info/Index) [25]. The genes annotated as CCR were retrieved by searching the keyword in genome annotation and hidden Markov model (HMM) profile using HMMER3.0 following previous work [26]. The cDNA sequence, coding sequence (CD), peptide sequence, and genomic DNA (gDNA) sequence of all CCR were downloaded from Ensemble Plants. The resulting candidate sequences were submitted to the NCBI, Pfam (Protein family, http://xfam.org/), and SMART (Simple Modular Architecture Research Tool, http://smart.emblheidelberg. de/) websites for validation and removing redundant sequences. The equivalent electric point (pI) and molecular weight (Mw) of each protein were obtained from the ExPASy website (http://web.expasy.org/compute_pi/). Each gene's subcellular location was predicted using Target1.1 Server, ChloroP1.1 Server, and SignalP5.0 [27]. The gene structure map was drawn based on the CDs and gDNA sequences of each gene to the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) to for intron-exon pattern and the number of introns for each gene.
2.2. Phylogenetic and conserved domain analyses
The amino acid sequences of 114 genes were aligned in the MEGA X software for sequence alignment [28]. The sequences were aligned in ClustalW to generate a mas file. A phylogenetic tree was constructed using the maximum likelihood estimation method in the MEGA X software. The boots-strap repetition number was set 1,000 times in the Poisson model to obtain the phylogenetic tree [28].
The amino acid sequences of wheat CCRs were aligned in the MEME website (http://meme-suite.org/) to predict the conserved domains by using the classic motif discovery mode. Novel conserved domains were filtered according to indicators such as E-value and sites for their detailed signature matches [29].
2.3. Chromosomal localization and the homologous CCRs Circos map
Each wheat chromosome's length and the start and end position of each CCR gene were from http://plants.ensembl.org/Triticum_aestivum/Info/Index. The information of CCR homoeologue pairs was extracted from the above website. The above homoeologue pairs information was uploaded into the Micro-Synteny View mode on the Tbtools website (https://github.com/CJ-Chen/TBtools/releases). The chromosomal localization of CCRs was created in the Advanced Circos tool on the Tbtools website.
2.4. Promoter element analysis
The 2.5 kb untranslated region (UTR) sequence upstream of the initiation codon (ATG) of each CCR was retrieved from the Ensemble Plants database to predict cis-acting regulatory elements (cis-elements). Cis-acting elements in the promoter sequences were predicted following previous work in the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/)[30]. The number of stress-related cis-acting elements predicted with each CCR gene was sorted into a table to draw the promoter element analysis map in the TB tools software (http://www.tbtools.com/).
2.5. Tissue and stress responsible expression characteristics of CCRs
Expression data of CCRs were downloaded from the wheat eFP website (http://bar.utoronto.ca/efp_wheat/cgi-bin/efpWeb.cgi) [31]. The expression information for each CCR in 51 tissue samples from spring wheat (Triticum aestivum) cv. Azhumaya [31] was transformed into a logarithm of the original TPM value into a combined table to draw a heat map in Graphpad Prism 8 (https://www.graphpad.com/scientific-software /prism/).
RNA-Seq data of abiotic and biotic stresses were downloaded from the wheat expression website (http://www.wheat-expression.com/). Expression data of 114 CCRs were extracted in the form of original TPM values and then converted to logarithmic values to draw heat maps in Graphpad Prism 8 (https://www.graphpad.com/). The images were colored in Adobe Illustrator (https://www.adobe.com/). The paired t-test was used to calculate further the significance between the control sample and the treated sample in each experiment.
2.6. Drought stress treatment analysis
Spring wheat (Triticum aestivum L.) cultivar Fielder was used in this study. The wheat seeds were sterilized with 75% ethanol for 5 minutes, then with 1% hydrochloric acid for 15 minutes, and finally washed three times with ddH2O. The treated seeds were spread on a crepe paper rinsed with tap water, kept in a refrigerator at 4°C overnight, and removed out to room temperature for 24h. Germinated seeds were sown in a black culture box filled with tap water. The culture box is enclosed in a growth chamber at 25°C with 80% humidity with a 16-hour light/8-hour dark-light cycle. Five seedlings at a leaf and a heart stage were treated with 20% PEG6000 for 1, 6, 12, 24 and 48 hours. The seedlings grown without PEG6000 were used as controls. The RNA was extracted from roots, stems, and leaves with Trizol reagent for reverse transcript HieffTM qPCR SYBRTM Green Master Mix (Shanghai Yeasen Biotechnology, Shanghai, China) in CFX 96-Realtime system (BIORAD, Singapore) according to the user manual. The corresponding primers were designed, and quantitative analyses were performed by qRT-PCR [32].
2.7. Determination of Lignin Content
In the lignin determination experiment, the first leaf of 4 wheat seedlings was collected and mixed as one biological replicate. Each sample has four biological replicates, each with five-technique repeats. Samples were frozen in the liquid nitrogen and ground into a fine powder in a tissue grinder at 55 Hz for 30 s for six times. The samples were washed with methanol three times following 1×PBS, 1% Triton X-100, 1 M NaCl, ddH2O, and acetone, three times each to remove soluble metabolites. The samples were kept overnight in a fume hood to let the acetone volatilize completely for alcohol-insoluble and protein-free cell walls (AIR) [33].
2.0±0.2 mg of each AIR sample was weighed into 2 mL centrifuge tubes in five biological repeats. The samples were mixed with 125μL 25% acetyl bromide (diluted with glacial acetic acid), treated at 70 °C for 35 min in a metal bath, and cooled to room temperature in a refrigerator at 4 °C. Then, 400 μL 2 M NaOH and 70 μL 0.5 M fresh hydroxylamine hydrochloride were sequentially added into the samples and tapped to mix well. The samples were adjusted to 2 ml by adding glacial acetic acid and centrifuged at 15,000rpm for 15 minutes. 200 μL of supernatant was carefully transferred into a 96-well UV detection plate to measure the absorbance with a microplate reader at a wavelength of 280 nm [34]. The lignin content measured by this method (the proportion coefficient of Gramineae is 17.75):
Lignin content (%) = [absorbance*constant volume (mL)]/[17.75*0.539*100% masses (mg)]
3.1. Phylogenetic relationship and conserved motifs of wheat CCRs
CCR family genes encoded in the wheat cultivar Chinese Spring (Triticum aestivum L.; ABD genome) were mined by searching publicly available databases and a hidden Markov model (HMM) search based on Pfam homologs. In total, 114 non-redundant CCRs were identified in the latest genome data of wheat. The putative wheat CCR proteins have a relatively narrow range in length, from 123 to 427 amino acids. Wheat CCR proteins have theoretical isoelectric points (pI) ranging from 5 to 9.5. A subcellular localization analysis revealed that 65 CCRs localized in the cytoplasm and 37 in the Golgi apparatus, while remaining in the chloroplast (5), mitochondrion (5), vacuole (4), plasma membrane (2, pI < 6), cell wall (2, pI > 8.7), and peroxisome (1) (Table S1).
To further evaluate the phylogenetic relationships, all the identified wheat CCRs were aligned to construct the wheat CCR gene family’s phylogenetic tree. In the phylogenetic analysis, wheat CCRs clustered into seven groups, with 6 to 25 genes in each subfamily (Fig. 1a). Chromosomal mapping revealed that group 7 CCRs were located on group 5 homeologoue chromosome, while group 6 CCRs group 7 homeologoue chromosomes of wheat (Table S1). Group 4 had an evolutionary relationship relatively distant from the other six groups, and most of them located on group 6 homeologoue chromosomes. More variations in the amino acid sequences in this group suggested that they might have diverged earlier in evolution or recently evolved with diverse biological functions.
We further analyzed conserved motifs in the CCRs based on their peptide sequences (Table S2). Three motifs were close to the enzyme's catalytic center and reported to be critical for the biological activity of CCR (Fig. 1b) [35]. The NWYCY domain works as the catalytic center, and the “WY” amino acids were also conserved in other species (Fig. 1b). The VTGA motif is associated with NAD(P)’ s binding and essential for CCR to catalyze the reaction (Fig. 1b). The RXXXXK motif is also recognized as a conserved motif notable for the binding specificity NADP (Fig. 1b). Besides the reported motifs, three new motifs were defined in wheat CCRs, which were also presented in rice CCRs (Fig. 1c). Further experimental support is needed to validate their potential roles in the biological function.
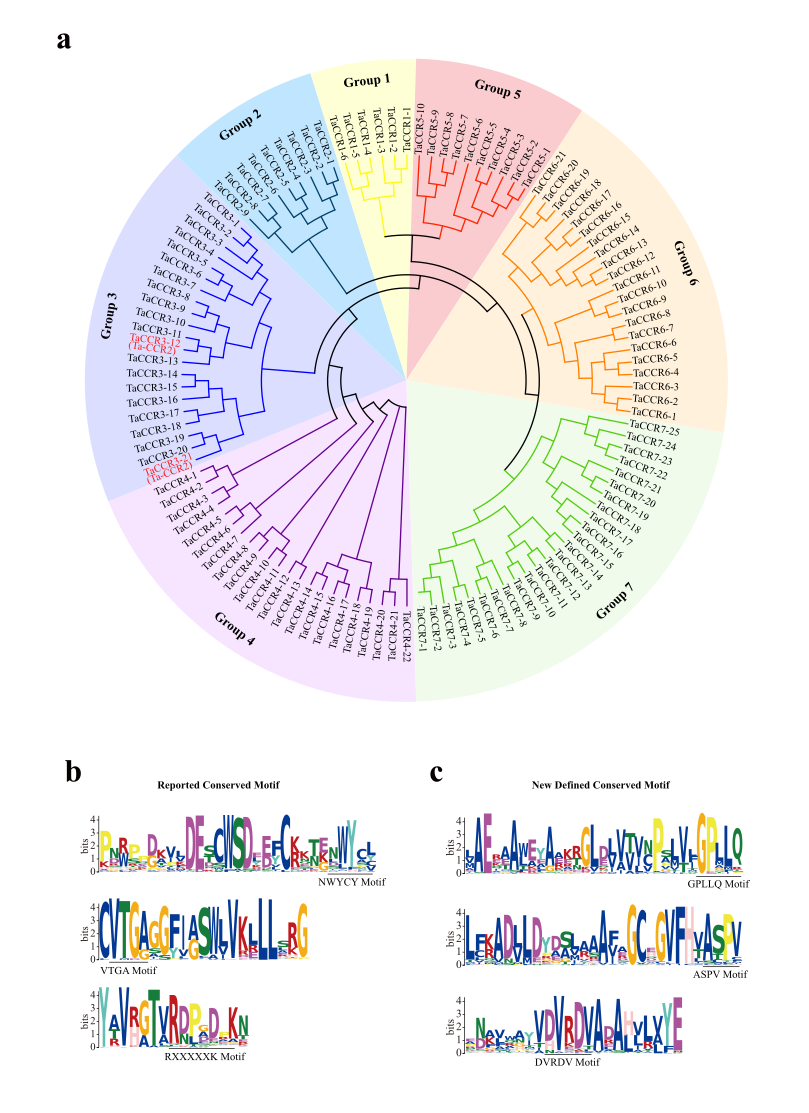
Fig. 1. Phylogenetic and conserved motif analysis of wheat CCR proteins. a, Phylogenetic relationship among wheat CCRs. b, Sequence constitution of reported NWYCY, VGTA, and RXXXXXK motifs in wheat CCRs. c Sequence constitution of three novel motifs in wheat CCRs.
3.2. Gene Structure analysis of wheat CCRs
Chromosome localization analysis showed that the 114 wheat CCRs distributed unevenly on the 21 chromosomes, with group 5 homeologoue chromosomes containing the most CCRs (Fig. 2). Two clusters of CCRs existed on chromosome 5 (5A, 5B and 5D) in large numbers as tandem repeats, and they had high homology in the phylogenetic tree (Table S1). These data suggested that the above genes could favor wheat’s environmental adaptability.
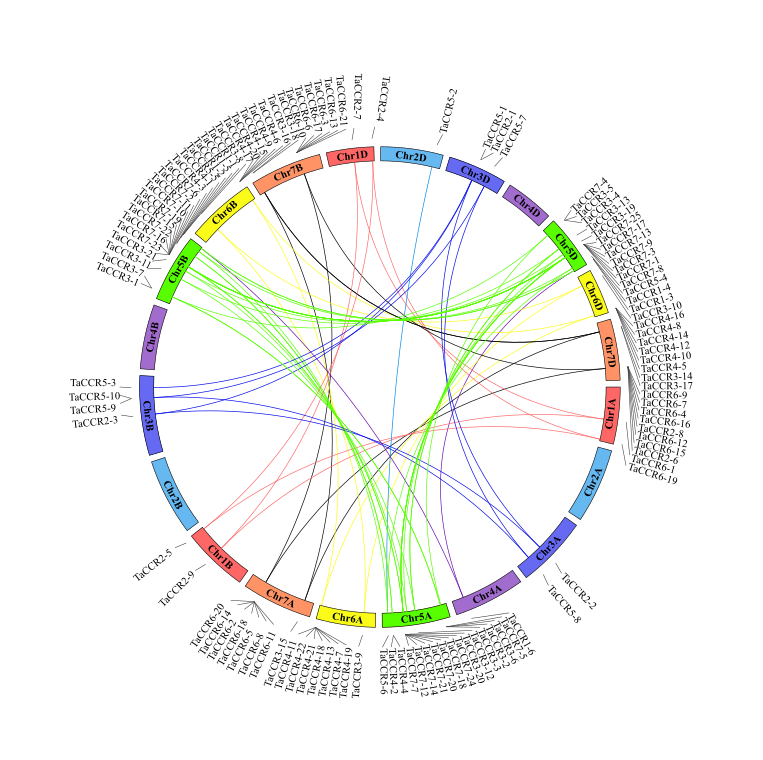
Fig. 2 Chromosomal localization and the homologous CCRs in wheat sub-genomes. Duplicated CCR genes in each homo-group are linked using lines with the corresponding color of the wheat chromosomes. The corresponding Traes ID for each CCR gene is listed in Table S1.
Cis-acting elements in the promoter sequences are the potential binding sites of the transcription factors that initiate transcription, which play a crucial role in regulating spatial and temporal gene expressions. We extracted the 2.5 kb upstream region of the translational start site of the 114 wheat CCRs to screen cis-regulatory elements. The CCR promoters contain ten potential cis-acting elements, including TC rich repeats (defense and stress responses), LTR (low-temperature response), TCA-element (salicylic acid, SA, response), ABRE (abscisic acid, ABA, response), CGTCA (methyl jasmonic acid, MeJA, response), TGACG (MeJA response), P-box (gibberellin, GA, response), GARE (GA response), TATC-box (GA response), MBS (MYB binding site in drought response), MBSI (MYB binding site in phenylpropanoid metabolism) and WUN (wound-responsible element) (Table S3). Based on a visual presentation of the cis-regulatory elements in CCRs’ promoter regions, ABA and MeJA responsible elements were presented in 97.5% and 95.6% of total CCR promoters, suggesting their high-abundance and wide-distributions (Table S3). The SA inducible TCA-element existed in 41.2% of the CCR promoters, consistent with the reported involvement of lignin biosynthesis in pathogenic responses. The drought and low-temperature inducible elements, MBS and LTR, were observed in 57.8% and 55.2% of CCR promoters, suggesting that the CCR family genes could play a role in wheat response to environmental stresses.
Gene structure (exon/intron) analysis provides clues to potential gene function, organization, evolution, and divergence patterns. Thus, we mapped the exon/intron organization of wheat CCRs. We found that 104 wheat CCRs have 3 to 5 introns (Fig. 3), which were highly conserved compared to the reported introns numbers in JAZ (0-7), ZIP (1-10), and MAPKK (1-7). For the CCRs with 1 and 2 introns, six out of 7 were among 13.64 to 21.37 in Mw, and are very likely truncated CCRs. The three truncated CCRs in group 3, TraesCS3B02G463900, TraesCS2D02G458800, and TraesCS3D02G221200, formed a small cluster, indicating that they have a common origin. However, the other truncated genes in group7, TraesCS6A02G380700, TraesCS6B02G420300, and TraesCS6A02G381300, have close homologs different from each other, suggesting that they were likely to be truncated as individual events during evolution.
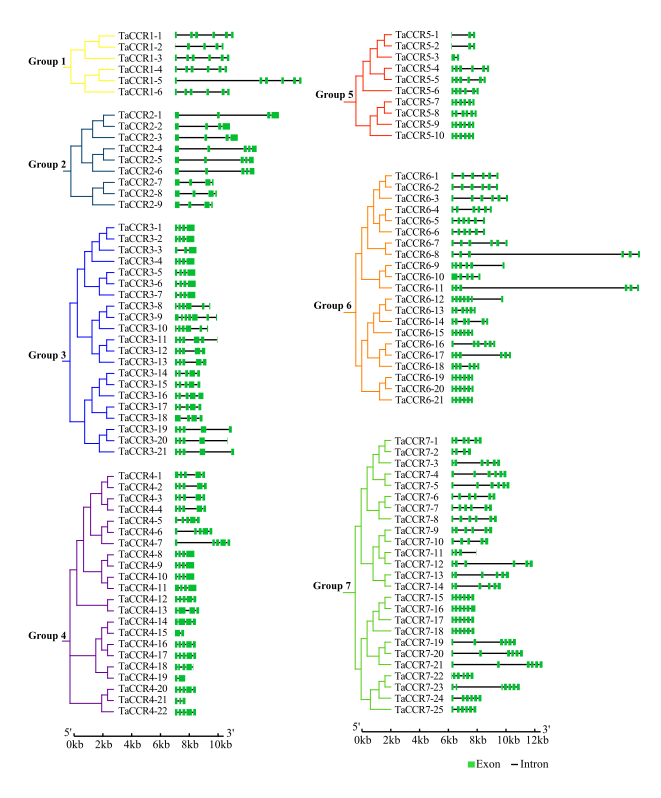
Fig. 3. Schematic gene structures of wheat CCR genes. The exons and introns are represented by green boxes and black horizontal lines, respectively.
3.3. The gene expression patterns of CCRs during wheat development
The phylogenetic relationship and motif patterns suggested that CCRs in different groups exhibit vast disparities in their binding motifs and potential affinities with substrates. The expression abundances among different tissues provide information for them to accommodate different physiological processes. We investigated the expression levels of wheat CCRs in over 50 wheat tissues at different developmental stages. Based on the gene expression patterns, groups 1-3, 5-7 had at least one pair of highly expressed homologs in all the tissues at the whole or most developmental stages (Fig. 4), which could play a dominant role amongst their group. Surprisingly, all the genes in group 4 had a relatively lower expression level, suggesting that they could have a minor biological function during wheat development, consistent with the significant divergence from other groups in the phylogenetic analysis.
Despite those above constitutively expressed genes, some CCRs only expressed in a particular tissue at a specific developmental stage (Table S4). The expression of TraesCS6B02G420000 and TraesCS6B02G420200 was very high in the root apical meristem at the three-leaf stage. Three genes in group 6, TraesCS7D02G392400, TraesCS7A02G398000, and TraesCS7B02G298900, were only expressed in the grains at soft dough to hard dough stages. The above expression pattern suggested that these genes are primarily involved in phenylpropanoid biosynthesis in the grain.
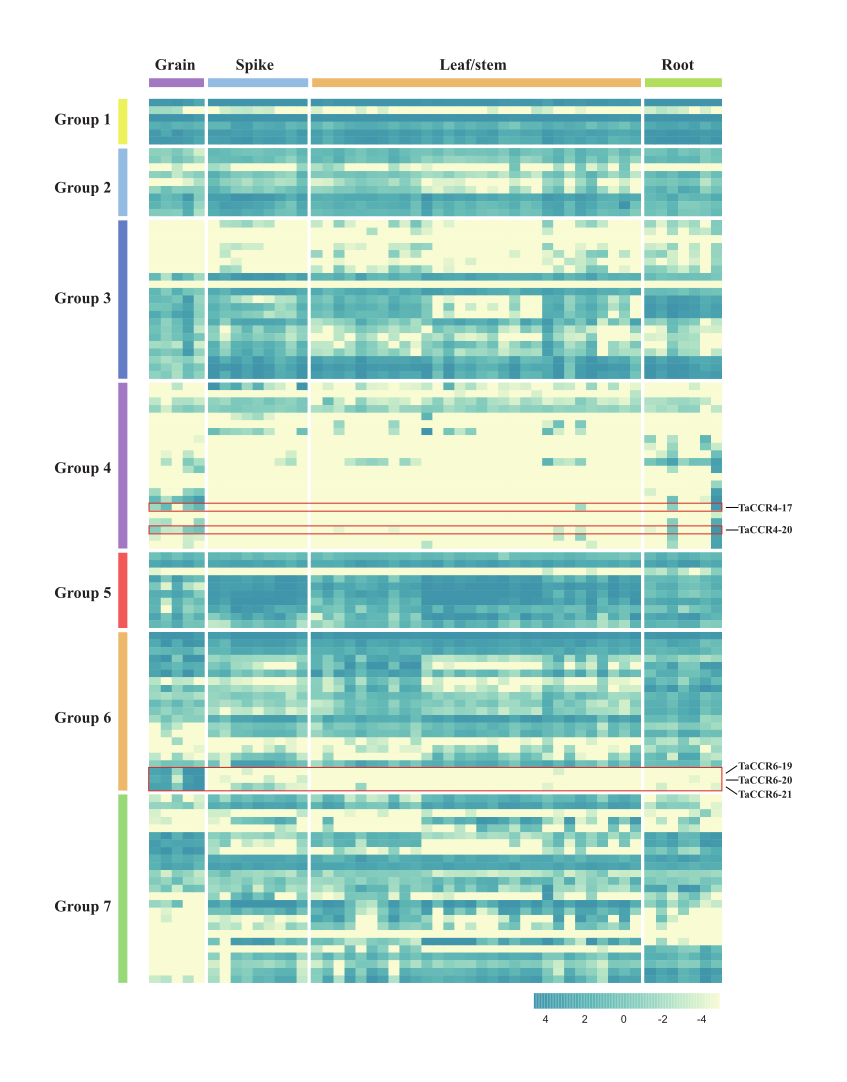
Fig. 4. Heat map of the expression profiles of CCRs in different tissues/developmental stages in wheat.
3.4. The gene expression patterns of CCRs during wheat stress responses
The phenylpropanoid pathway, including lignin and flavonoids, is often co-upregulated in stress responses [36]. We set to analyze the expression of CCRs for candidates involved in defense responses, including harsh environmental stresses on seedlings and divesting fungal diseases, e.g., biotrophic Pst on leaf and semi-biotrophic Fusarium graminearum on spikelet (Table S5).
We first analyzed the expression of CCRs in the wheat-pathogen interacting system. Upon inoculation with Pst 87/66, the overall expression level increased to around two folds in 1-day post-inoculation (dpi), declined from 2~5 days, and increased again in the later stage, e.g., 1.5-folds of mock at 11dpi, (Fig. 5a, Table S6). Further analyses showed that groups 2, 6, and 7 contributed to the early induction, while group 1 was dominant in the later induction (Fig. 5b). Upon inoculation with Fusarium graminearum, CCRs were induced in anthesis-stage spikelets (only using palea, lemma, and rachis) from 24 to 48 hours (Table S6). Group 3 was induced at the highest level by the infection of Fusarium graminearum (Fig. 5d). The induction patterns suggested that different CCR subgroups could play different roles to biotrophic (Pst) and necrotrophic (Fusarium graminearum) fungi at different inoculation stages.
Next, we analyzed the expression of CCRs during wheat responses to harsh environmental stresses. There was no significant difference from control at the whole genome level in response to drought, heat, and PEG6000 (students’ pair-wise t-test value, P > 0.05) (Fig. 5e). A minor decline of some CCRs was also observed in a combined treatment of heat and drought treatment (Fig. 5f). In a recent meta-analysis across plant species, CCR1 presented as a conserved gene repressed by heat [37]. Therefore, based on these transcriptome data, it would be logical to predict that CCRs could play a minor role in wheat response to environmental stresses.
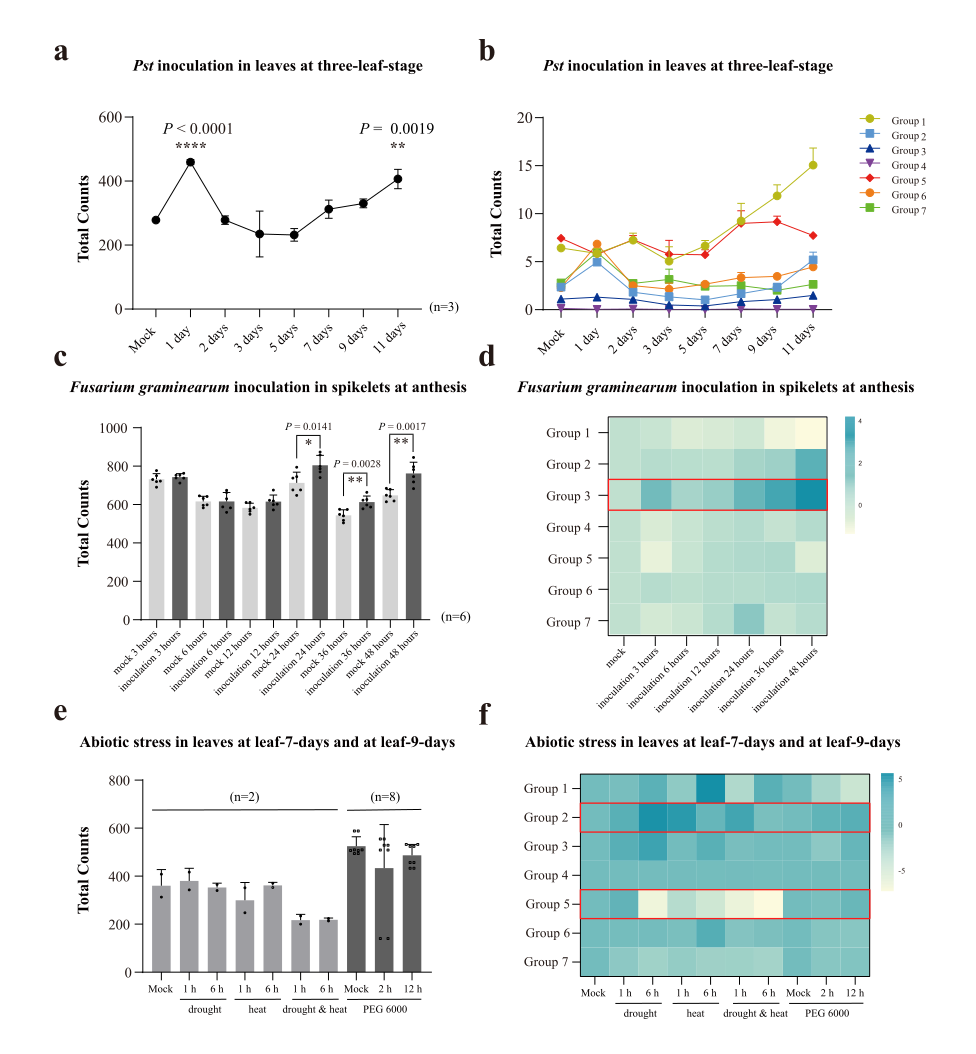
Fig. 5. The expression profiles of CCRs in response to abiotic and biotic stresses. a, The overall expression of CCRs in response to Pst at different inoculation stages. b, The expression of CCR subgroups in response to Pst at different inoculation stages. c, The overall expression of CCRs in spikes inoculated by Fusarium graminearum at different stages. d, The expression of CCR subgroups in response to Fusarium graminearum at different stages. e, The overall expression of CCRs in heat and drought treatments. f, The expression of CCR subgroups in response to abiotic stresses at different stages. Pair-wise students’ t-test P-value, * < 0.05, ** < 0.01, **** < 0.0001. Bar = ±s.d.
3.5. Lignin content and CCR expression changes in wheat drought response
Lignification represents an essential evolution in adaptation to terrestrial environments for the plants and is closely related to plant drought resistance [13]. In the above transcriptome analysis, wheat CCRs did not significantly increase, which led to a logical inconsistency between CCRs’ gene expression and known concepts.
First, we quantified the lignin contents in drought-stressed seedling leaves assimilated by Polyethylene glycol treatment. The short-term drought treatments, e.g., 12 and 24 hours, did not significantly affect the lignin contents of wheat seedling leaves. The variance analysis showed a significant increase in lignin content after a drought treatment for 48 hours (Fig. 6a, Supplementary Fig. 3). The above change of lignin content in the drought-treatment suggested that lignin biosynthesis and the related genes’ expression represented metabolism changes later than primary signaling pathways in wheat leaves. We predicted that the treatment duration time of 1 to 6 hours could not be sufficient for CCRs to increase.
To test the above hypothesis, we conducted the same drought treatment and analyzed the expression of CCRs at different time points. The expression of TraesCS1B02G209700 increased at 1hour, while TraesCS3B02G380500 and TraesCS5B02G386500 decreased at 6hour upon lignin treatment in the roots (Fig. 6b). In the leaves, all the three CCRs, TraesCS1B02G209700, TraesCS3B02G380500, and TraesCS5B02G486500, significantly increased at 12 H (Fig. 6c).
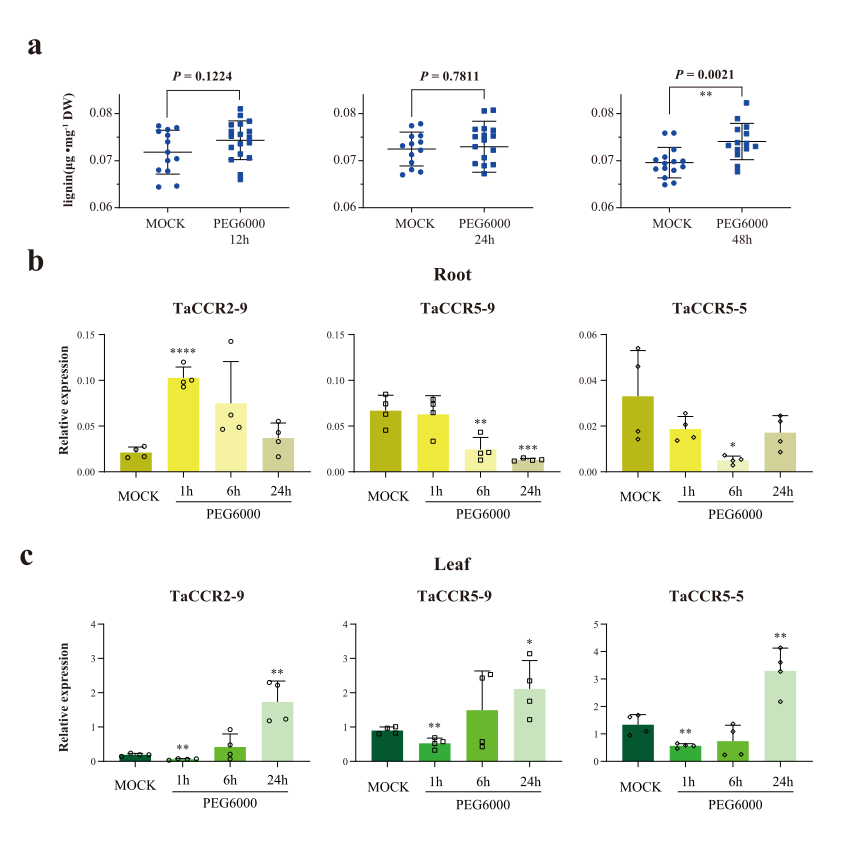
Fig. 6. The change of lignin content and expression profiles of CCRs in drought-treated wheat seedlings. a, The change of lignin contents in seedlings. The data represented the average of four biological repeats, each with five-technique repeats. b-c, The relative expression levels of representative CCRs in drought treatment in the root (b) and leaf (c). Students’ t-test P-value, * < 0.05, ** < 0.01, *** < 0.001, **** < 0.0001. n = 4.
CCR proteins belong to a conserved gene family in the plant kingdom to confer a vital role during plant growth and development and protect the plant from abiotic and biotic stress by synthesizing lignin to adapt to the terrestrial environment. These enzymes' functions have been widely studied, including model and crop plants, e.g., Arabidopsis and rice. However, fewer efforts have been devoted to investigating the function of CCR gene family in wheat. In this study, we identified and characterized the CCR gene family in wheat at the whole-genome level.
A total of 114 putative wheat CCRs belonging to 7 CCR subfamilies were obtained through a genome-wide search approach. The number of CCRs is far beyond those in rice and Populus tomentosa, which have 33 and 11 CCRs, respectively [38]. Gene duplication is an essential mechanism for acquiring new genes and creating genetic novelty during evolution in organisms [39]. CCRs existed in chromosomes 5 (5A, 5B, and 5D) in higher number (Fig. 2), representing tandem duplication, one of the two crucial gene amplification ways. Moreover, the significant increase of CCR gene number in wheat over model plants also reflects whole-genome duplication due to hexaploid (AABBDD) formed by ancient hybridizations among the ancestral wheat species [40].
The CCR gene family’s functional studies were conducted in many plants to investigate the gene structure, protein structure, biochemical characters, and stress responses [41, 42]. Switchgrass (Panicum virgatum) contains a diverse gene family of CCR, which are phylogenetically distinct, differentially expressed, and have different biochemical properties [43]. Sorghum CCRs showed evolutionary conservation of the functional domains in a structure and phylogeny analysis but were differentially expressed in a drought treatment [44]. Previous biochemical studies provided useful information for the role of TaCCR1 and TaCCR2 in stem development [27, 45]. However, further investigation of CCRs at the whole genome level is necessary for a full view image.
The 114 wheat CCRs were distributed in 7 distinct subgroups based on their phylogenetic relationships. The majority of the CCRs fall into one of the three intron-exon models reported in rice, and each model has a similar intron length among different members in them [46]. Most of the CCRs shared the reported basic conserved motifs, e.g., NWYCY exists in 102 CCRs, as the characteristic motif to recognize CCR in the plants. Besides the reported motifs, three novel conserved motifs present in different subfamilies, indicating that they could contribute to different subfamilies’ specific biochemical characters.
Large-scale prediction of promoter sequences and their contributing cis-acting elements are crucial for improving our fundamental understanding of gene regulation [47]. Here, wheat CCR promoters contain a wide array of cis-acting elements ranging from plant hormone (MeJA, ABA, SA, and GA) response to stress responses (cold, drought, and defense). Deleting an MBSIIG MYB cis-element in EgCCR promoters blocked the DNA-protein complex’s formation in EMSA -electrophoretic mobility shift assay experiments and the transcriptional activation of the EgCCR promoter. A correlation was presented between some cis-elements in CCR promoters and the stress-induced gene expression in this work. Therefore, the corresponding cis-elements could involve regulating CCRs for the adaption of wheat to environmental stresses.
Lignin is critical for plants’ adaption to terrestrial environments during evolution [13]. A positive relationship between lignin and drought could enhance water transport and prevent water loss, therefore promoted drought tolerance in plants [48]. In a drought stress treatment simulated by 10% PEG6000 treatment, SiMYB56 overexpression in rice accumulated more lignin and had significantly higher root length, plant height, and biomass, which represented a drought resistance phenotype [49]. Here, three CCRs significantly increased at 24 hours upon drought treatment, consistent with the increase of lignin content at 48 hours. The above expression changes lead to a conclusion different from the transcriptome data (Fig. 5e) and earlier reports [37], in which samples were collected at 1 to 6 hours. This difference suggested different sampling times for the analyses of master regulators (early) or enzymes catalyzing biochemical reactions (late).
Post-translational modifications (PTM) also have significant impacts on the biological functions of enzymes. An auxin-responsive Kelch-domain-containing F-box protein, OsFBK1, binds OsCCR and leads to its degradation through the 26S proteasome pathway to control lignification in the development of rice anthers and roots [50]. OsRac1, one of the Rac/Rop family of small GTPases, interacts with and activates OsCCR1 to accumulate lignin and increase reactive oxygen species production during defense responses in rice [21]. Therefore, CCRs with known PTMs and the groups with different induction patterns among stress responses should have priority in future functional studies.
Wheat CCRs formed seven subfamilies with different distributions, gene structures, and functional motifs. The expression variations among different CCRs during development and stresses suggested that certain CCRs’ subfamilies could play essential roles in corresponding biological processes. The evidence suggests that lignin biosynthesis displays a significant change in drought stress.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Financial support
This research is supported by the National Natural Science Foundation of China (31772146 and 31972350).
S. Bekele, S. Melinda, B. Hans-Joachim, D. Etienne, R. Mathew, M. Geoffrey, Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security, Food Security 5(3) (2013).
View ArticleS. Daryanto, L. Wang, P.A. Jacinthe, Global Synthesis of Drought Effects on Maize and Wheat Production, PLoS One 11(5) (2016) e0156362. PMid:27223810
View Article PubMed/NCBIM. Lamaoui, M. Jemo, R. Datla, F. Bekkaoui, Heat and Drought Stresses in Crops and Approaches for Their Mitigation, Front Chem 6 (2018) 26. PMid:29520357
View Article PubMed/NCBIS. Tounsi, K. Feki, D. Hmidi, K. Masmoudi, F. Brini, Salt stress reveals differential physiological, biochemical and molecular responses in T. monococcum and T. durum wheat genotypes, Physiol Mol Biol Plants 23(3) (2017) 517-528. PMid:28878491
View Article PubMed/NCBIX. Li, J. Cai, F. Liu, T. Dai, W. Cao, D. Jiang, Cold priming drives the sub-cellular antioxidant systems to protect photosynthetic electron transport against subsequent low temperature stress in winter wheat, Plant Physiol Biochem 82 (2014) 34-43. PMid:24887010
View Article PubMed/NCBIM. Djanaguiraman, D.L. Boyle, R. Welti, S.V.K. Jagadish, P.V.V. Prasad, Decreased photosynthetic rate under high temperature in wheat is due to lipid desaturation, oxidation, acylation, and damage of organelles, BMC Plant Biol 18(1) (2018) 55. PMid:29621997
View Article PubMed/NCBIM. Abid, Y. Shao, S. Liu, F. Wang, J. Gao, D. Jiang, Z. Tian, T. Dai, Pre-drought priming sustains grain development under post-anthesis drought stress by regulating the growth hormones in winter wheat (Triticum aestivum L.), Planta 246(3) (2017) 509-524. PMid:28526982
View Article PubMed/NCBIA. Jighly, M. Alagu, F. Makdis, M. Singh, S. Singh, L.C. Emebiri, F.C. Ogbonnaya, Genomic regions conferring resistance to multiple fungal pathogens in synthetic hexaploid wheat, Molecular Breeding 36(9) (2016) 127.
View ArticleJ. Sthapit, E.E. Gbur, G. Brown-Guedira, D.S. Marshall, E.A. Milus, Characterization of Resistance to Stripe Rust in Contemporary Cultivars and Lines of Winter Wheat from the Eastern United States, Plant Dis 96(5) (2012) 737-745. PMid:30727527
View Article PubMed/NCBIJ.M. Beddow, P.G. Pardey, Y. Chai, T.M. Hurley, D.J. Kriticos, H.J. Braun, R.F. Park, W.S. Cuddy, T. Yonow, Research investment implications of shifts in the global geography of wheat stripe rust, Nat Plants 1 (2015) 15132. PMid:27251389
View Article PubMed/NCBIK. Kazan, D.M. Gardiner, J.M. Manners, On the trail of a cereal killer: recent advances in Fusarium graminearum pathogenomics and host resistance, Mol Plant Pathol 13(4) (2012) 399-413. PMid:22098555
View Article PubMed/NCBIJ.C. Moura, C.A. Bonine, J. de Oliveira Fernandes Viana, M.C. Dornelas, P. Mazzafera, Abiotic and biotic stresses and changes in the lignin content and composition in plants, J Integr Plant Biol 52(4) (2010) 360-76. JIPB892 PMid:20377698
View Article PubMed/NCBIQ. Liu, L. Luo, L. Zheng, Lignins: Biosynthesis and Biological Functions in Plants, Int J Mol Sci 19(2) (2018). PMid:29364145
View Article PubMed/NCBIJ.K. Weng, C. Chapple, The origin and evolution of lignin biosynthesis, New Phytol 187(2) (2010) 273-85. PMid:20642725
View Article PubMed/NCBIR. Vanholme, B. De Meester, J. Ralph, W. Boerjan, Lignin biosynthesis and its integration into metabolism, Curr Opin Biotechnol 56 (2019) 230-239. PMid:30913460
View Article PubMed/NCBIQ.-H. Ma, H.-H. Zhu, M.-Y. Qiao, Contribution of both lignin content and sinapyl monomer to disease resistance in tobacco, Plant Pathology 67(3) (2018) 642-650.
View ArticleT. Vogt, Phenylpropanoid biosynthesis, Mol Plant 3(1) (2010) 2-20. PMid:20035037
View Article PubMed/NCBIL. Jones, A.R. Ennos, S.R. Turner, Cloning and characterization of irregular xylem4 (irx4): a severely lignin-deficient mutant of Arabidopsis, Plant J 26(2) (2001) 205-16. PMid:11389761
View Article PubMed/NCBIR. Vanholme, B. Demedts, K. Morreel, J. Ralph, W. Boerjan, Lignin biosynthesis and structure, Plant Physiol 153(3) (2010) 895-905. PMid:20472751
View Article PubMed/NCBIA.M. Boudet, S. Hawkins, S. Rochange, The polymorphism of the genes/enzymes involved in the last two reductive steps of monolignol synthesis: what is the functional significance?, C R Biol 327(9-10) (2004) 837-45. PMid:15587075
View Article PubMed/NCBIT. Kawasaki, H. Koita, T. Nakatsubo, K. Hasegawa, K. Wakabayashi, H. Takahashi, K. Umemura, T. Umezawa, K. Shimamoto, Cinnamoyl-CoA reductase, a key enzyme in lignin biosynthesis, is an effector of small GTPase Rac in defense signaling in rice, Proc Natl Acad Sci U S A 103(1) (2006) 230-5. PMid:16380417
View Article PubMed/NCBIH.L. Park, S.H. Bhoo, M. Kwon, S.W. Lee, M.H. Cho, Biochemical and Expression Analyses of the Rice Cinnamoyl-CoA Reductase Gene Family, Front Plant Sci 8 (2017) 2099. PMid:29312373
View Article PubMed/NCBIA. Yu, J. Zhao, Z. Wang, K. Cheng, P. Zhang, G. Tian, X. Liu, E. Guo, Y. Du, Y. Wang, Transcriptome and metabolite analysis reveal the drought tolerance of foxtail millet significantly correlated with phenylpropanoids-related pathways during germination process under PEG stress, BMC Plant Biol 20(1) (2020) 274. PMid:32539796
View Article PubMed/NCBIS. Srivastava, R.K. Vishwakarma, Y.A. Arafat, S.K. Gupta, B.M. Khan, Abiotic stress induces change in Cinnamoyl CoA Reductase (CCR) protein abundance and lignin deposition in developing seedlings of Leucaena leucocephala, Physiol Mol Biol Plants 21(2) (2015) 197-205. PMid:25931776
View Article PubMed/NCBIE. Pennisi, Detailed genome maps paths to better wheat, Science 361(6403) (2018) 635. PMid:30115790
View Article PubMed/NCBIX. Xu, B. Yan, Y. Zhao, F. Wang, X. Zhao, L. He, J. Xu, C. Zhao, Characterization and expression analysis of GPAT gene family in maize, Canadian Journal of Plant Science 99(5) (2019) 577-588.
View ArticleO. Emanuelsson, S. Brunak, G. von Heijne, H. Nielsen, Locating proteins in the cell using TargetP, SignalP and related tools, Nat Protoc 2(4) (2007) 953-71. PMid:17446895
View Article PubMed/NCBIS. Kumar, G. Stecher, M. Li, C. Knyaz, K. Tamura, MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms, Mol Biol Evol 35(6) (2018) 1547-1549. PMid:29722887
View Article PubMed/NCBIS. Hunter, R. Apweiler, T.K. Attwood, A. Bairoch, A. Bateman, D. Binns, P. Bork, U. Das, L. Daugherty, L. Duquenne, R.D. Finn, J. Gough, D. Haft, N. Hulo, D. Kahn, E. Kelly, A. Laugraud, I. Letunic, D. Lonsdale, R. Lopez, M. Madera, J. Maslen, C. McAnulla, J. McDowall, J. Mistry, A. Mitchell, N. Mulder, D. Natale, C. Orengo, A.F. Quinn, J.D. Selengut, C.J. Sigrist, M. Thimma, P.D. Thomas, F. Valentin, D. Wilson, C.H. Wu, C. Yeats, InterPro: the integrative protein signature database, Nucleic Acids Res 37(Database issue) (2009) D211-5. PMid:18940856
View Article PubMed/NCBIL. Guo, J. Gao, J.A. Teixeira da Silva, X. Yu, Bioinformatics analysis of the auxin response factor gene family in Prunus persica, Canadian Journal of Plant Science 99(2) (2018) 232-242.
View ArticleR.H. Ramirez-Gonzalez, P. Borrill, D. Lang, S.A. Harrington, J. Brinton, L. Venturini, M. Davey, J. Jacobs, F. van Ex, A. Pasha, Y. Khedikar, S.J. Robinson, A.T. Cory, T. Florio, L. Concia, C. Juery, H. Schoonbeek, B. Steuernagel, D. Xiang, C.J. Ridout, B. Chalhoub, K.F.X. Mayer, M. Benhamed, D. Latrasse, A. Bendahmane, B.B.H. Wulff, R. Appels, V. Tiwari, R. Datla, F. Choulet, C.J. Pozniak, N.J. Provart, A.G. Sharpe, E. Paux, M. Spannagl, A. Brautigam, C. Uauy, The transcriptional landscape of polyploid wheat, Science 361(6403) (2018). PMid:30115782
View Article PubMed/NCBIS. Wang, Q.P. Li, J. Wang, Y. Yan, G.L. Zhang, Y. Yan, H. Zhang, J. Wu, F. Chen, X. Wang, Z. Kang, J. Dubcovsky, J.Y. Gou, YR36/WKS1-Mediated Phosphorylation of PsbO, an Extrinsic Member of Photosystem II, Inhibits Photosynthesis and Confers Stripe Rust Resistance in Wheat, Mol Plant 12(12) (2019) 1639-1650. PMid:31622682
View Article PubMed/NCBIJ.Y. Gou, X.H. Yu, C.J. Liu, A hydroxycinnamoyltransferase responsible for synthesizing suberin aromatics in Arabidopsis, Proc Natl Acad Sci U S A 106(44) (2009) 18855-60. PMid:19846769
View Article PubMed/NCBIC.E. Foster, T.M. Martin, M. Pauly, Comprehensive compositional analysis of plant cell walls (Lignocellulosic biomass) part I: lignin, J Vis Exp (37) (2010).
View ArticleH. Pan, R. Zhou, G.V. Louie, J.K. Muhlemann, E.K. Bomati, M.E. Bowman, N. Dudareva, R.A. Dixon, J.P. Noel, X. Wang, Structural studies of cinnamoyl-CoA reductase and cinnamyl-alcohol dehydrogenase, key enzymes of monolignol biosynthesis, Plant Cell 26(9) (2014) 3709-27. PMid:25217505
View Article PubMed/NCBIR.A. Dixon, N.L. Paiva, Stress-Induced Phenylpropanoid Metabolism, Plant Cell 7(7) (1995) 1085-1097. PMid:12242399
View Article PubMed/NCBII. Balti, J. Benny, A. Perrone, T. Caruso, D. Abdallah, A. Salhi-Hannachi, F. Martinelli, Identification of conserved genes linked to responses to abiotic stresses in leaves among different plant species, Funct Plant Biol (2020). PMid:32727652
View Article PubMed/NCBIN. Chao, N. Li, Q. Qi, S. Li, T. Lv, X.N. Jiang, Y. Gai, Characterization of the cinnamoyl-CoA reductase (CCR) gene family in Populus tomentosa reveals the enzymatic active sites and evolution of CCR, Planta 245(1) (2017) 61-75. PMid:27580618
View Article PubMed/NCBIS. Magadum, U. Banerjee, P. Murugan, D. Gangapur, R. Ravikesavan, Gene duplication as a major force in evolution, 92 (2013) 155-61. PMid:23640422
View Article PubMed/NCBIT. Marcussen, S.R. Sandve, L. Heier, M. Spannagl, M. Pfeifer, K.S. Jakobsen, B.B. Wulff, B. Steuernagel, K.F. Mayer, O.A. Olsen, Ancient hybridizations among the ancestral genomes of bread wheat, Science 345(6194) (2014) 1250092.
J. Raes, A. Rohde, J.H. Christensen, Y. Van de Peer, W. Boerjan, Genome-wide characterization of the lignification toolbox in Arabidopsis, Plant Physiol 133(3) (2003) 1051-71. PMid:14612585
View Article PubMed/NCBIS.A. Sattler, A.M. Walker, W. Vermerris, S.E. Sattler, C. Kang, Structural and Biochemical Characterization of Cinnamoyl-CoA Reductases, Plant Physiol 173(2) (2017) 1031-1044. PMid:27956488
View Article PubMed/NCBIL.L. Escamilla-Trevino, H. Shen, S.R. Uppalapati, T. Ray, Y. Tang, T. Hernandez, Y. Yin, Y. Xu, R.A. Dixon, Switchgrass (Panicum virgatum) possesses a divergent family of cinnamoyl CoA reductases with distinct biochemical properties, New Phytol 185(1) (2010) 143-55. PMid:19761442
View Article PubMed/NCBIJ. Li, F. Fan, L. Wang, Q. Zhan, P. Wu, J. Du, X. Yang, Y. Liu, Cloning and expression analysis of cinnamoyl-CoA reductase (CCR) genes in sorghum, PeerJ 4 (2016) e2005. PMid:27231650
View Article PubMed/NCBIQ.H. Ma, B. Tian, Biochemical characterization of a cinnamoyl-CoA reductase from wheat, Biol Chem 386(6) (2005) 553-60. PMid:16006242
View Article PubMed/NCBIA. Barakat, N.B. Yassin, J.S. Park, A. Choi, J. Herr, J.E. Carlson, Comparative and phylogenomic analyses of cinnamoyl-CoA reductase and cinnamoyl-CoA-reductase-like gene family in land plants, Plant Sci 181(3) (2011) 249-57. PMid:21763535
View Article PubMed/NCBIC.M. Hernandez-Garcia, J.J. Finer, Identification and validation of promoters and cis-acting regulatory elements, Plant Sci 217-218 (2014) 109-19. PMid:24467902
View Article PubMed/NCBIW. Liu, Y. Jiang, C. Wang, L. Zhao, Y. Jin, Q. Xing, M. Li, T. Lv, H. Qi, Lignin synthesized by CmCAD2 and CmCAD3 in oriental melon (Cucumis melo L.) seedlings contributes to drought tolerance, Plant Mol Biol 103(6) (2020) 689-704. PMid:32472480
View Article PubMed/NCBIW. Xu, W. Tang, C. Wang, L. Ge, J. Sun, X. Qi, Z. He, Y. Zhou, J. Chen, Z. Xu, Y.Z. Ma, M. Chen, SiMYB56 Confers Drought Stress Tolerance in Transgenic Rice by Regulating Lignin Biosynthesis and ABA Signaling Pathway, Front Plant Sci 11 (2020) 785. PMid:32625221
View Article PubMed/NCBIP. Borah, J.P. Khurana, The OsFBK1 E3 Ligase Subunit Affects Anther and Root Secondary Cell Wall Thickenings by Mediating Turnover of a Cinnamoyl-CoA Reductase, Plant Physiol 176(3) (2018) 2148-2165. PMid:29295941
View Article PubMed/NCBI