Email: weihuaiwu2002@163.com (W.W.); yikexian@126.com (K.Y.)
Genome-wide analysis of defensin-like genes in Coffea arabica
Corresponding Author
Affiliation
Xing Huang 1, Thomas Gbokie Jr. 2, Baohui Liu 2, Weihuai Wu 1,*, and Kexian Yi 1,*
1 Environment and Plant Protection Institute, Chinese Academy of Tropical Agricultural Sciences, Haikou 571101, China; hxalong@gmail.com (X.H.); weihuaiwu2002@163.com (W.W.); yikexian@126.com (K.Y.)
2 College of Plant Protection, Nanjing Agricultural University, Nanjing 210095, China; gbokiejr@gmail.com (T.G.); baohuiliu2019@163.com (B.L.)
Article Reviewed By:
Y P Cai(ypcaiah@163.com)
Fei Liu(liuhuijuedui@163.com)
Zhaoen Yang(yangzhaoen0925@126.com)
Bing Zhang(bingzhang@yzu.edu.cn)
Citation
Xing Huang,Weihuai Wu , Kexian Yi Genome-wide analysis of defensin-like genes in Coffea arabica(2019)SDRP Journal of Plant Science 3(2) p:159-164
Abstract
Coffee leaf rust disease caused by Hemileia vastatrix, is currently a very serious problem in coffee production and resistant varieties are the most appropriate and sustainable mitigation strategy against the disease. But the knowledge for both constitutive and inducible immune system is still limited in C. arabica. In this present study, we conducted genome-wide analysis of defensin-like genes, an important peptide family of plant constitutive immunity, in C. arabica genome. Eighteen defensin-like genes were obtained by TBLASTn method and further clustered into two groups by phylogenetic analysis. Nucleotide sequence alignment revealed insertion/deletion mutations of duplicated defensin-like genes in C. arabica. Besides, structure modeling apparently indicated two types of protein structures among the 18 defensin-like genes. This study provides an overview of defensin-like genes in C. arabica and will serve as a guideline for future study.
Keywords: Coffea arabica; defensin-like genes; phylogeny; protein structure
Introduction
Coffee is a well appreciated beverage crop in the world and a cash crop of huge socioeconomic importance for many smallholder farmers in developing countries. Coffea arabica and Coffea canephora are main commercially produced species ofo coffee [1]. Leaf rust disease caused by a biotrophic fungus Hemileia vastatrix (Uredinales: Pucciniaceae) is a very serious phytosanitory problems in the production of this crop [2]. Fungicide is an efficient way to control the disease, however, associated the production cost and concerns over the adverse effects of misapplications on the quality of coffee drinks and the environment make resistant varieties being the most appropriate and sustainable management strategy against the disease [3]. A series of resistant varieties were released, including Oeiras-MG 6851 that originated from the cross Caturra Vermelho (CIFC 19/1) and Hibrido de Timor (CIFC 832/1) [4]. But the resistance has already been broken by race XXXIII of H. vastatrix [5]. Thus, it is really of great importance to study the interaction and co-evolution between coffee and H. vastatrix. Coffee-H. vastatrix rust interactions are governed by the gene-for-gene relationship [6]. To date, transcriptome and proteome methods has been used to find resistance genes [7,8]. In spite of the reported dynamics of H. vastatrix effector candidate genes (HvECs), the existing knowledge of resistance genes (SH1-SH9) is still limited [9]. The plant immune system consists of both constitutive and inducible mechanisms [10]. Defensin is an important peptide family of plant constitutive immunity [11]. Some exogenous defensin genes have been successfully transformed in crops and improved the disease resistance of transgenic lines [12-14]. Although resistance genes (SH1-SH9) has already been used for coffee breeding, there are however few reports on coffee defensin. Here, we present the genome-wide analysis of defensin-like genes in C. arabica by the released bioinformation on Phytozome. The results provide an overview for defensin-like genes in C. arabica and will serve as a guideline for future study.
Materials & Methods
2.1. Sequence retrieval
The seed sequences of conserved domain in defensin proteins were downloaded from Pfam database under the accession of PF00304 [15]. These sequences were used as query to search against C. arabica genome database from Phytozome [16] by using TBLASTN with a cut off Expected value (E-value) of 10−5 [17]. The information of these genes on scaffold position were also obtained.
2.2. Phylogenetic analysis
A neighbor-joining (NJ) phylogenetic tree was constructed for defensin proteins using the Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0 [18]. Bootstrap values from 1000 trials were used for constructing the most parsimonious tree.
2.3. Intron analysis
Obtained genomic and coding sequences of defensin-like genes in C. arabica were employed for intron analysis. The exon/intron structures were analyzed using the online Gene Structure Display Server (GSDS) with coding sequences and genomic sequences [19].
2.4. Motif analysis
Amino acid sequences were aligned using Clustal X [20]. The alignment was employed for motif analysis in DNAMAN software (Lynnon Corporation, VauDREUIL-DORION QC Canada) [21].
2.5. Structure modeling
Translated protein sequences of defensin-like genes in C. arabica were used for structure modeling by PHYRE2 Protein Fold Recognition Server [22].
Results
3.1. Identification of defensin-like genes in C. arabica
After removing the redundant sequences, we obtained 18 defensin-like genes in the genome database of C. arabica line UCG-17 (Table 1). Their genomic sequence lengths were from 476-1065 base pairs (bp). The coding sequence length of these genes ranged from 225 to 321 bp, encoding peptides of 74-106 amino acids (aa). Their scaffold locations were also obtained when sequence retrieval, which indicated that 5 gene clusters, were formed.
3.2. Phylogenetic and intron analysis
The 18 protein sequences were clustered into 2 groups (Figure 1A). Group I contained 16 sequences, which is far more than Group II. Group I was further divided into 2 subgroups with each containing 8 sequences. Intron/exon for each defensin-like gene was analyzed according to their genomic and coding sequences (Figure 1B). All the genes contained one (1) intron, respectively. And the intron lengths were from 239-828.
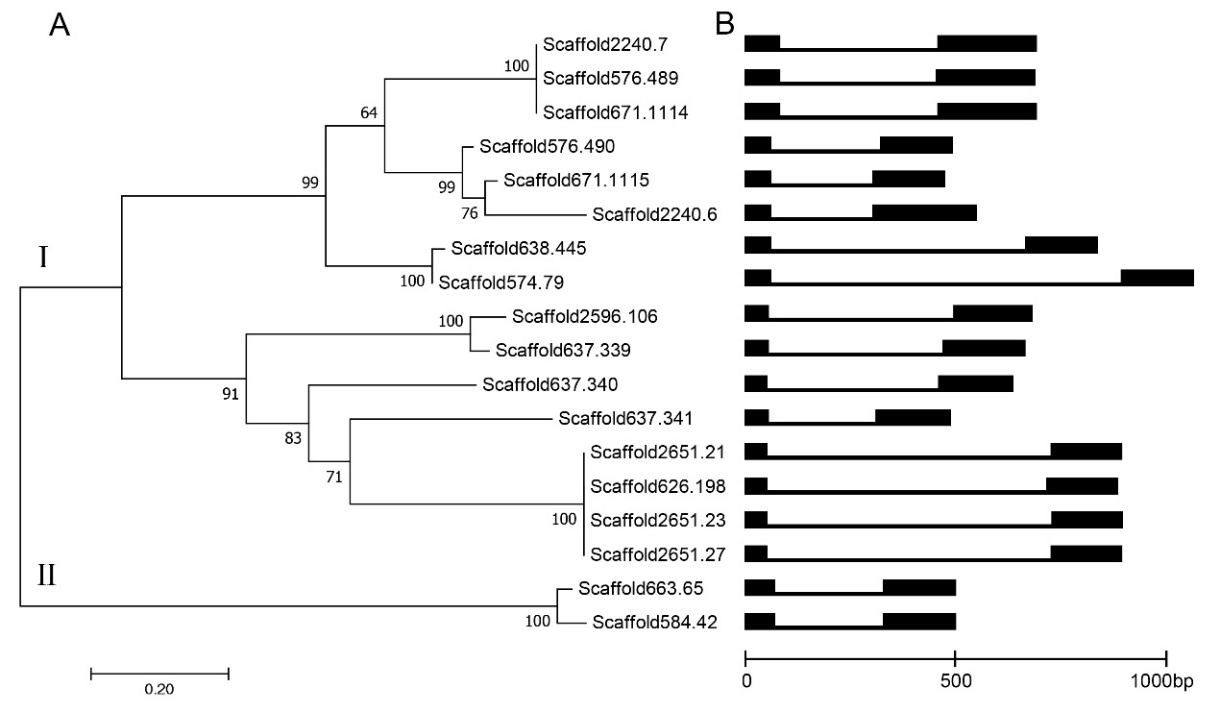
Figure 1. Phylogenetic analysis of defensin proteins in C. arabica (A). Bootstrap values were from 1000 trials. Intron/exon configurations of defensin-like genes in C. arabica (B). Introns and exons drawn to scale with the full encoding regions of their respective genes with boxes indicating the exons, and lines representing introns.
Table 1. Defensin-like gene family in C. arabica.
|
Accession |
Genomic (bp) |
CDS (bp) |
Protein (aa) |
Intron Length (bp) |
Position |
Strand |
|
Scaffold574.79 |
1065 |
237 |
78 |
828 |
Scaffold574:831842-832906 |
+ |
|
Scaffold576.489 |
690 |
321 |
106 |
369 |
Scaffold576:3566049-3566738 |
+ |
|
Scaffold576.490 |
495 |
237 |
78 |
258 |
Scaffold576:3568764-3569258 |
+ |
|
Scaffold584.42 |
502 |
246 |
81 |
256 |
Scaffold584:412397-412898 |
+ |
|
Scaffold626.198 |
884 |
225 |
74 |
659 |
Scaffold626:2859425-2860308 |
- |
|
Scaffold637.339 |
666 |
255 |
84 |
411 |
Scaffold637:2549770-2550435 |
+ |
|
Scaffold637.340 |
638 |
234 |
77 |
404 |
Scaffold637:2552861-2553498 |
+ |
|
Scaffold637.341 |
491 |
237 |
78 |
254 |
Scaffold637:2554488-2554978 |
+ |
|
Scaffold638.445 |
839 |
237 |
78 |
602 |
Scaffold638:5246186-5247024 |
+ |
|
Scaffold663.65 |
502 |
246 |
81 |
256 |
Scaffold663:766014-766653 |
+ |
|
Scaffold671.1114 |
693 |
321 |
106 |
372 |
Scaffold671:9167520-9168212 |
+ |
|
Scaffold671.1115 |
476 |
237 |
78 |
239 |
Scaffold671:9169969-9170444 |
+ |
|
Scaffold2240.6 |
551 |
312 |
103 |
239 |
Scaffold2240:30833-31383 |
- |
|
Scaffold2240.7 |
693 |
321 |
106 |
372 |
Scaffold2240:33139-33831 |
- |
|
Scaffold2596.106 |
677 |
246 |
81 |
431 |
Scaffold2596:734206-734882 |
+ |
|
Scaffold2651.21 |
895 |
225 |
74 |
670 |
Scaffold2651:351557-352451 |
- |
|
Scaffold2651.23 |
897 |
225 |
74 |
672 |
Scaffold2651:367736-368632 |
- |
|
Scaffold2651.27 |
895 |
225 |
74 |
670 |
Scaffold2651:402056-402950 |
- |
3.3. Motif analysis and structure modeling
Plant defensin family contains the gamma thionin domain (PF00304), which was also observed in C. arabica defensins (Figure 2). The alignment result indicated that two gene pairs shared the same protein sequences, i.e. Scaffold671.1114/2240.7 and Scaffold2651.21/2651.23/2651.27/626.198. We further analyzed their nucleotide sequences and found that the genomic sequences of Scaffold671.1114 and Scaffold2240.7 were the same, also between Scaffold2651.21 and Scaffold2651.27. Scaffold2651.23 shared the same sequence in coding region with Scaffold2651.21 and Scaffold2651.27, but an insertion mutation in intron region (Figure 3). Scaffold626.198 had two deletion mutations in intron region and two synonymous mutation in coding region. Besides, there was frameshift mutation caused by single nucleotide deletion in Saffold2240.6 (Figure 4).
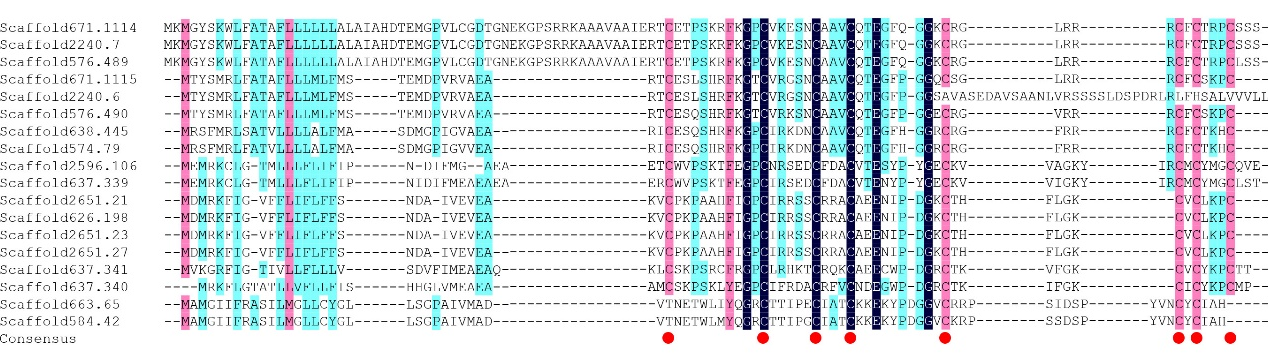
Figure 2. Alignment of defensin proteins in C. arabica. Conserved cysteine residues were highlighted with red dot.

Figure 3. Mutation configurations (genomic sequence) of Scaffold2651.23 and Scaffold626.198 compared with Scaffold2651.21/2651.27. The numbers represent the length of the sequence.
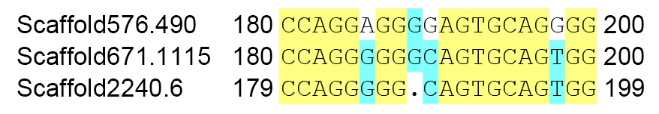
Figure 4. Frameshift mutation caused by single nucleotide deletion in coding sequence of Scaffold2240.6. The numbers represent the length of the sequence.
We further constructed the protein structure models by PHYRE2 Protein Fold Recognition Server. All the 16 proteins in Group I matched the d1gpsa model of PHYRE2 (Figure 5A), while the 2 proteins in Group II matched the d1n4na model from PHYRE2 (Figure 5B).
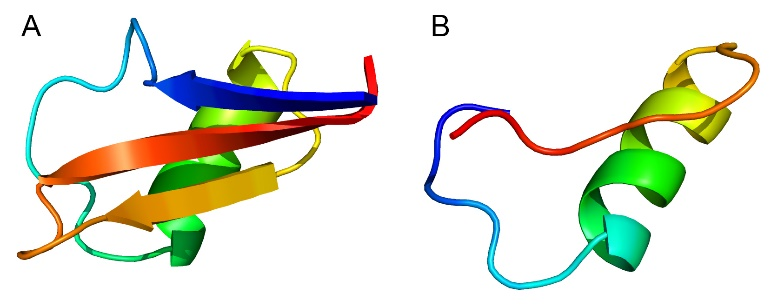
Figure 5. Structure models of proteins in Group I (A) and Group II (B) based on d1gpsa and d1n4na models from PHYRE2, respectively.
Discussion
In this study, we successfully identified 18 defensin-like genes in C. arabica genome, which is far less than those characterized in Arabidopsis genome [23]. Besides, a recent study revealed a similar scale of defensin-like genes in sorghum (10), maize (20), rice (12) and brachypodium (9) genomes [24]. Such a result might however not be adequate enough to provide understanding on the evolution pattern of defensin-like genes. According to phylogenetic analysis and alignment conducted, we found that two gene pairs (Scaffold671.1114/2240.7 and Scaffold2651.21/2651.27) shared the same sequence, respectively. Scaffold671.1114 and Scaffold2240.7 could very likely be allelic genes. The C. arabica genome was not assembled as high quality as C. canephora, which further emphases the difficulty for genome assembly for tetraploid [25]. More further research works are still needed to confirm the relation between Scaffold671.1114 and Scaffold2240.7, despite the assertion that Scaffold2651.21 and Scaffold2651.27 are duplicated genes. Because they are from the same scaffold, the 5 gene clusters should be also generated by whole genome duplication [26]. Duplicated genes are commonly accompanied with expansion of gene family and mutation of gene function. Our result provided an example for gene mutation at early stage (Figure 3). Besides, the frameshift mutation in Scaffold2240.6 is believed to cause the change in the tail part of the protein sequence (Figure 2; Figure 4). This mutation might cause an irreversible change of the protein function. It is worth noting that the 16 defensins categorized in Group I matched d1gpsa model in PHYRE2, and their structure models were consistent with reported defensins [11]. Moreover, Group II defensins only had partial structure compared with Group I, which might weaken their antifungal activity. Recombinant expression analysis for these defensins in C. arabica would be necessary and efficient to characterize their functions in future studies [27].
Acknowledgement
We would like to thank Margit Laimer from University of Natural Resources and Life Sciences, BOKU-VIBT (Vienna 1190, Austria) for her thorough suggestions on the experimental design
Author Contributions: Conceptualization: X.H., W.W. and K.Y.; Formal analysis: X.H.; Funding acquisition: W.W. and K.Y.; Investigation: X.H., T.G. and B.L.; Supervision: K.Y.; Writing—original draft: X.H.; Writing—review and revise: T.G.
Funding: This study was funded by National Key R&D Program of China (2018YFD0201100), FAO/IAEA Collaborative Research Project (20380), International Exchange and Cooperation Project of the Ministry of Agriculture (SYZ2019), Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (1630042017021, 1630042019030), Special Funds for Efficient Tropical Agriculture Development of Hainan Province (UF37721).
References
Davis, A.P.; Tosh, J.; Ruch, N.; Fay, M.F. Growing coffee: Psilanthus (Rubiaceae) subsumed on the basis of molecular and morphological data; implications for the size, morphology, distribution and evolutionary history of Coffea. Bot. J. Linn. Soc. 2011, 167, 357-377.
View ArticleZambolim L. Current status and management of coffee leaf rust in Brazil. Trop Plant Pathol 2016, 41, 1-8.
View ArticleVárzea, V.M.P.; Marques, D.V. Resistance, population variability of Hemileia vastatrix vs. coffee durable. In Durable resistance to coffee leaf rust; Zambolim, L.; Zambolim, E.; Várzea, V.M.P., Eds.; Publisher: UFV Press, Viçosa, Brazil, 2005; pp. 53-74.
Pereira, A.A.; Zambolim, L.; Chaves, G.M.; Sakiyama, N.S. Cultivar de café resistente à Ferrugem: Oeiras-MG 6851. Rev. Ceres. 2000, 46, 121-124.
Capucho, A.S.; Zambolim, E.M.; Freitas, R.L.; Haddad, F.; Caixeta, E.T.; Zambolim, L. Identification of race XXXIII of Hemileia vastatrix on Coffea arabica Catimor derivatives in Brazil. Australas Plant Dis. Notes 2012, 7, 189-191.
View ArticleFlor, H.H. Inherence of pathogenicity in Melampsora lini. Phytopathol. 1942, 32, 653-669.
Guerra-Guimarães, L.; Tenente, R.; Pinheiro, C.; Chaves, I.; Silva Mdo, C.; Cardoso, F.M.; Planchon, S.; Barros, D.R.; Renaut, J.; Ricardo, C.P. Proteomic analysis of apoplastic fluid of Coffea arabica leaves highlights novel biomarkers for resistance against Hemileia vastatrix. Front. Plant Sci. 2015, 6, 478. PMid:26175744
View Article PubMed/NCBIFlorez, J.C.; Mofatto, L.S.; Freitas‑Lopes, R.L.; Ferreira, S.S.; Zambolim, E.M.; Carazzolle, M.F.; Zambolim, L.; Caixeta, E.T. High throughput transcriptome analysis of coffee reveals prehaustorial resistance in response to Hemileia vastatrix infection. Plant Mol. Biol. 2017, 95, 1-17. PMid:29094279
View Article PubMed/NCBIMaia, T.; Badel, J.L.; Marin-Ramirez, G.; Rocha, C.M.; Fernandes, M.B.; Silva, J.C.F.; Azevedo-Junior, G.M.; Brommonschenkel, S.H. The Hemileia vastatrix effector HvEC6 suppresses bacterial blight symptoms in coffee genotypes with the SH1 rust resistance gene. New Phytol. 2017, 213, 1315-1329. PMid:27918080
View Article PubMed/NCBIDoehlemann, G.; Hemetsberger, C. Apoplastic immunity and its suppression by filamentous plant pathogens. New Phytol. 2013, 198, 1001-1016. PMid:23594392
View Article PubMed/NCBIThomma, B.P.; Cammue, B.P.; Thevissen, K. Plant defensins. Planta 2002, 216, 193-202. PMid:12447532
View Article PubMed/NCBISwathi, A.T.; Divya, K.; Jami, S.K.; Kirti, P.B. Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep. 2008, 27, 1777-1786. PMid:18758784
View Article PubMed/NCBINtui, V.O.; Thirukkumaran, G.; Azadi, P.; Khan, R.S.; Nakamura, I.; Mii, M. Stable integration and expression of wasabi defensin gene in "Egusi" melon (Colocynthis citrullus L.) confers resistance to Fusarium wilt and Alternaria leaf spot. Plant Cell Rep. 2010, 29, 943-954. PMid:20552202
View Article PubMed/NCBIGaspar, Y.M.; Mckenna, J.A.; Mcginness, B.S.; Hinch, J.; Poon, S.; Connelly, A.A.; Anderson, M.A.; Heath, R.L. Field resistance to Fusarium oxysporum and Verticillium dahliae in transgenic cotton expressing the plant defensin NaD1. J. Exp. Bot. 2014, 65, 1541. PMid:24502957
View Article PubMed/NCBIEl-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427-D432. PMid:30357350
View Article PubMed/NCBIGoodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178-D1186. PMid:22110026
View Article PubMed/NCBIAltschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389-3402. PMid:9254694
View Article PubMed/NCBIKumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870-1874. PMid:27004904
View Article PubMed/NCBIHu, B.; Jin, J.; Guo, A.Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: an upgraded gene feature visualization server. Bioinform. 2015, 31, 1296-1297. PMid:25504850
View Article PubMed/NCBIJeanmougin, F.; Thompson, J.D.; Gouy, M.; Higgins, D.G.; Gibson, T.J. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 1998, 23, 403-405. 01285-7
View ArticleDNAMAN-Bioinformatics Solutions. www.lynnon.com (accessed on 20-03-2019).
Bennett-Lovsey, R.M.; Herbert, A.D.; Sternberg, M.J.; Kelley, L.A. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins 2008, 70, 611-625. PMid:17876813
View Article PubMed/NCBISilverstein, K.A.T.; Graham, M.A.; Paape, T.D.; VandenBosch, K.A. Genome Organization of More Than 300 Defensin-Like Genes in Arabidopsis. Plant Physiol. 2005, 138, 600-610. PMid:15955924
View Article PubMed/NCBIWu, J.; Jin, X.; Zhao, Y.; Dong, Q.; Jiang, H.; Ma, Q. Evolution of the defensin-like gene family in grass genomes. J. Genet. 2016, 95, 53. PMid:27019432
View Article PubMed/NCBIDenoeud, F.; Carreteropaulet, L.; Dereeper, A.; Droc, G.; Guyot, R.; Pietrella, M.; Zheng, C.; Alberti, A.; Anthony, F.; Aprea, G.; Aury, J.M.; Bento, P.; Bernard, M. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Sci. 2014, 345, 1181-1184. PMid:25190796
View Article PubMed/NCBIJaillon, O.; Aury, J.M.; Wincker, P. "Changing by dou-bling", the impact of whole genome duplications in the evolution of eukaryotes. C. R. Biol. 2009, 332, 241-253. PMid:19281955
View Article PubMed/NCBIPicart, P.; Pirttilä, A.M.; Raventos, D.; Kristensen, H.H.; Sahl, H.G. Identification of defensin-encoding genes of Picea glauca: characterization of PgD5, a conserved spruce defensin with strong antifungal activity. BMC Plant Biol. 2012, 12, 180. PMid:23035776
View Article PubMed/NCBI
