Zhilong Wang
Email: wangzhilong@nbcc.cn
© 2019 Sift Desk Journals. All Rights Reserved
Zhilong Wang
Email: wangzhilong@nbcc.cn
Wen Li 1, Jiaying Wang2, Xintong Jiang1, Qianhui Zhu1, Yueqiu He 1, Tao Fu 1, and Zhilong Wang 1,*
1 Key Laboratory of Plant Development and City College of Vocational Technology·Utilization of Ningbo, Ningbo Zhejiang 315199, China; 2 Ningbo Academy of Inspection and Quarantine, Ningbo 315012, China
E-mail address: liwen547249317@126.com (Wen Li), 691062809@qq.com (Jiaying Wang), 18848406@qq.com (Yueqiu He), 819969334@qq.com (Tao Fu), wangzhilong@nbcc.cn (Zhilin Wang)
Cruz-Martin(mileidy@ibp.co.cu)
Liu JQ(liujieqing@hqu.edu.cn)
Frikha-Gargouri O(olfa.frikhagargouri@cbs.rnrt.tn)
Lu CL(luyixiao-0707@163.com)
Wen Li, Jiaying Wang, Xintong Jiang, Qianhui Zhu, Yueqiu He ,Tao Fu ,Zhilong Wang, Identification and Characterization of a Pseudomonas mosselii strain and its Antibacterial Function against Agrobacterium tumefaciens (2020)Journal of Plant Science 4(2) :223-231
Agrobacterium tumefaciens is a gram negative bacterial that can infect a range of plants and result in root crown gall. A total number of 10328 bacterial strains were isolated from rhizosphere of cherry tree. One strain of LWB10 showed clear inhibition zone around the bacterial colony in YEB media inoculated with A. tumefaciens C58. Morphological, physiological, and biochemical characterization indicated that LWB10 belongs to member of the genus Pseudomonas. Results from the high-throughput matrix-assisted laser desorption/ionization biotypersmart system indicated that this strain had a score value of 2.247 relative to Pseudomonas mosselii. Also, phylogenetic analysis based on 16S rRNA gene sequence showed that strain LWB10 shared the highest similarity with Pseudomonas mosselii CIP 105259T. The antagonist strains also exhibit well in growth inhibition of other five A. tumefaciens strains. Coinoculation of LWB10 and plant pathogenic strain of A. tumefaciens CFCC1369 showed strongly inhibition of tumor formation in tomato stems. All the results demonstrated that the isolated strain is P. mosselii LWB10 and its antibacterial ability to A. tumefaciens may offer new way for management of crown gall disease in the future.
Keywords: Agrobacterium tumefaciens, Pseudomonas mosselii, antagonist, biological control
Agrobacterium tumefaciens is a kind of soil-born pathogen that causes crown gall in roots [1, 2]. Crown gall had found to occur on nearly 40 economically important plants, such as Amygdalus persica L., Cerasus spp., Malus domestica, Pyrus spp. and Vitis vinifera L. [3, 4]. Cerasus sp. is widely cultivated in China for its beautiful blooming flowers. But recently, crown gall disease spread fast in cherry yard and people found that it is hard to control this disease.
As one of the destructive soil-born pathogen, A. tumefaciens will produce tumors in root and rhizome upon infection and thus affect water and nutrient absorption from roots. The disease developed slowly and disease symptoms will not be recognized until plants get weak or die [5]. Chemical control is the most common strategy to control the disease. But no symptoms appear in the early stage of infection and it is too late for the chemical control when we notice the disease. It will cause significant economically losses, when the pathogen once established in the field.
Therefore, researchers have been trying to find ways on dealing with the pathogen before they infect the roots. Among all the disease control methods, biological control is a kind of strategy that can protect plants away from pathogenic microorganisms. The most common way is to control of soil-born plant pathogens in the rhizosphere with bacteria. Bacterial genera studied for the control of crown gall disease include Pseudomonas, Bacillus, Serratia and Agrobaterium. The most successfully used antagonist strain is A. radiobacter K84 [6-10]. This strain was isolated in Australia, and it has an inhibitory action on most pathogenic A. tumefaciens containing the nopaline Ti plasmid (biotype Ⅰand Ⅱ), although some strains of A. tumefaciens biotype Ⅰ and Ⅱ became resistant to K84 agrocin [11]. Dipping seeds or roots into a K84 suspension prior to planting can efficiently prevent crown gall formation in the field on the roots of roses [12] and cherry [13]. Moreover, a novel B. methylotrophicus strain 39b found to stop the growth of A. tumefaciens C58 and B6. Mass spectrometry analysis revealed surfactins as the active principle that acting against Agrobacterium strains [14].
The genus Pseudomonas is a group of Gram-negative bacteria that are rod shaped, aerobic, non-spore-forming and motile [15]. Pseudomonas strains are capable to survive in diverse niches, ranging from terrestrial and aquatic environments to tissues of eukaryotic hosts. Many members of this genus displayed remarkable physiological and metabolic activity against different pathogens [16-19]. The highly precision metabolic system and several secondary metabolites, including phenazines, pyrrolnitrin, pyoluteorin and lipopeptides help Pseudomonas against other bacteria and fungi [20].
In this study, a total number of 10328 bacterial strains were isolated from rhizosphere of cherry tree to test their antibiotic ability for the control of A. tumefaciens. One strain named LWB10 showed strong antibiotic activity to A. tumefaciens C58 in vitro. Zone of inhibition and co-culture assay demonstrated that P. mosselii LWB10 significantly inhibited the growth of several pathogenic A. tumefaciens strains. When coinoculated LWB10 with two pathogenic A. tumefaciens strains in tomato, a significant decrease of tumor observed. Then we used a series of methods to identify this strain. Results from morphological, physiological, biochemical characterization, molecular identification and MALDI-TOF analysis indicated that this strain belonged to P. mosselii. Moreover, the antibiotic components could secrete outside the cell. Collectively, our results revealed that P. mosselii LWB10 is a promising biocontrol agent for inhibiting the growth of A. tumefaciens.
2.1. Sample collection
We collected soil samples from the root area of a cherry tree in Ningbo, China. The entire collected samples were isolated immediately for the microbiological experiments in the laboratory.
2.2. Isolation of antibacterial strains
The soil samples were serially diluted and inoculated on LB agar media (1% tryptone, 0.5% yeast extract, 0.5% NaCl, 1% agar-agar per 1 liter) at 28 ºC for 48 h. After that we isolated and ordered all the clear monoclones in individual tubes with LB liquid media in a shaker under the same temperature. All the bacteria were potential antibiotic agent for A. tumefaciens C58. Then A. tumefaciens was grown at 28 ºC in YEB medium (0.5% tryptone, 0.1% yeast extract, 0.5% beef extract, 0.5% sucrose and 0.05% MgSO4 per 1 liter) until the optical density at 600 nm (OD600) reached about 0.8. Inhibition zone assays conducted to reveal the antibiotic ability of isolated strains against C58. Top agar medium was prepared according to the following steps. There are three layers in the top agar medium. In the first layer, we poured 15 mL YEB solid medium in sterilized petrol dish. The second layer is the mixture of 1 ml A. tumefaciens C58 (density at OD600 about 1.0) and 10 mL YEB solid medium (cold till 60 ºC). Nine sterilized paper discs (0.6 cm in diameter) placed on the surface of the plate to form the third layer. Then 10 μL of each candidate antibacterial strains at OD600 of 1.0 were loaded on the paper discs[21, 22]. The zone of inhibition of bacterial growth appeared after 24 h incubation at 28 ºC. Other strains of A. tumefaciens used in this study including GV3101, LBA4404, EHA105, ACCC19197 (ordered from Agricultural Culture Collection of China) and CFCC1369 (ordered from China Forest Culture Collection Center). The antibiotic ability of candidate strains against to other A. tumefaciens strains were tested with the same procedure.
2.3. Morphological, physiological and biochemical characterization
Transmission electron microscopes (TEM) used for examining the cell morphology of the candidate strain after cells grown for 12 h on YEB medium. Gram staining conducted according to the procedure described by Murray [23]. Bacteria was cultured on YEB medium at different temperature of 4, 16, 22, 28, 42 ºC for 2-4 d to detect cell growth. Biochemical and physiological characterization examined using non-fermenting microorganism identification tube according to the manufactures’ instructions (HuanKai Microbial, China).
2.4. Identification bacteria by MALDI Biotyper platform
First, candidate bacteria grew on the YEB agar plate for 24 h at 28 ºC. Then cells collected and washed twice with sterilized water for protein extraction according to the ethanol/formic acid extraction method. Four technical replicates were spotted onto a MALDI target plate and analyzed by MALDI-TOF MS (Matrix-assisted laser desorption/lionization time of flight mass spectrometry) according to the manufacturer’s instructions [24]. Later, the spectra were loaded into Biotype software The Biotyper-derived scores obtained according to the comparision against the MSP database library.
2.5. Phylogenetic analysis
Genomic DNA extracted using the TIANamp Bacteria DNA Kit (Cat. No DP302, TIANGEN BIOTECH (BEIJING) CO. LTD) according to the manufacturer’s instructions. The 16S rRNA gene amplified by PCR using the universal primer pair: F8 (5’-AGAGTTTGATCCTGGCTCAG-3’)/ R1492 (5’-ACGGCTACCTTGTTACGACTT-3’). Fragment sequenced and sequence similarity was determined using BLAST server of NCBI. The available sequences from NCBI database which showing >99% similarity retrieved by BLAST N program available at NCBI server (www.ncbi.nlm.nih.gov). Phylogenetic tree based on 16S rRNA genes reconstructed using MEGA of version 7.0 [25]. Bootstrap analysis based on 1000 replications used to estimate the confidence level of tree topologies.
2.6. The antibiotic activity in vivo
Tomato plants were used as model to evaluate the antibiotic ability of isolated strains. Seeds were sown in individual pot for 1-2 month in growth chambers (at 25 ± 2 ºC, 65% relative humility, and 12/12 h light/darkness photoperiod). Plants main stems with 6-8 true leaves were pin-prick inoculated with pathogenic A. tumefaciens strains of CFCC1369 or the antibiotic strain alone or the mixture of antibiotic strain with A. tumefaciens. The procedures were conducted as following: first, dipped the sterilized absorbent cotton in the bacterial solution (1×108 cfu mL-1). Then attached the main stems with absorbent cotton after pin-prick and covered with plastic wrap for 2 d in growth chamber. After that the absorbent cotton were detached from main stems and disease symptoms were evaluated at 30-60 days post inoculation (dpi) by calculating the size of tumor in inoculated sites. Each treatment conducted with 15 individual replications.
3.1. LWB10 shows strong inhibitory activity against C58
A total number of 10328 culturable bacterial colonies obtained from cherry trees root area. Among them, nine isolates demonstrated various degrees of inhibitory activities against C58 growth (zone of inhibition area >10 mm) after two days of cocultivation. Notably, a bacterial isolate (termed LWB10) showed strong inhibition zone to A. tumefaciens C58 than other bacterial strain (LWB11) (Fig. 1a). The diameter of inhibition zone was > 20 mm after 2 d. Further study revealed that LWB10 could produce diffusible yellowish pigments on the YEB agar medium after incubation for 1-5 days (Fig. 1b). Moreover, we coculture LWB10 with C58 for several days and found that LWB10 showed strong inhibition zone to A. tumefaciens C58. The inhibition zone had a diameter of 26, 40 and 62 mm after coculture with A. tumefaciens C58 after 2, 4 and 6 days (Fig. 1c, in 90 mm petri dish). These results indicate that LWB10 effectively inhibited the growth of A. tumefaciens C58.
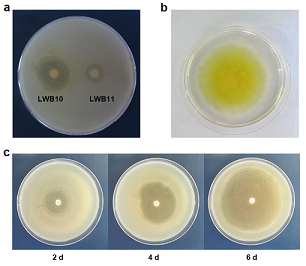
Fig. 1 Strain LWB10 shows strong inhibitory activity against C58. (a), LWB10 showed bigger inhibition zone than LWB11; (b) LWB10 produces diffusible yellowish pigments on YEB agar plate after 1 day of incubation, picture was taken at 5 dpi; (c) Zone of inhibition test showed that after incubation for 2, 4 and 6 days, the inhibition zone of LWB10 to C58 was 26, 40 and 62 mm, respectively.
3.2. Molecular identification of LWB10
Blast analysis demonstrated that LWB10 closely related to the genus Pseudomonas. The phylogenies of LWB10 and other 32 members of the genus Pseudomonas were determined by using the neighbor-joining method in the program MEGA7.0 [25]. As showed in Fig. 2, the tree has two big branches. One branch contains Pseudomonas pachastrellae CCUG 46540T, Pseudomonas azotoformans DSM 18862T, Pseudomonas composti CCUG 59231T and Pseudomonas punonensis CECT 8089T. Strain LWB101 clustered in another branch with Pseudomonas mosselii CIP 105259T. The sequence similarity of LWB10 to Pseudomonas mosselii CIP 105259T is as high as 98%. All these results indicated that LWB10 belongs to Pseudomonas mosselii.

Fig. 2 Phylogenetic tree analysis of LWB10 based on the nucleotide sequence of 16S rRNA gene. The tree was generated by the neighbor joining (NJ) method. Bootstrap probability values of above 50% are indicated at branch-points.
3.3. Morphological, physiological and biochemical characterization
Results from morphological, physiological and biochemical tests showed that strain LWB10 was determined to be G-, non-spore-forming rods (0.5-0.8 μm in wide and 1.5-1.8 μm in long) with polar flagella, motile as shown in Fig. 3a and 3b. Cell colonies on blood agar medium are circular and smooth. Colonies have a 2 mm diameter length after growth for 24 h (Fig. 3c). Bacteria could not growth under 4 and 42 ºC. The biochemical characteristics of strain LWB10 listed in table 1. According to the biochemical characteristics, we concluded the bacteria was P. mosselii.
Table 1. The biochemical characteristics of strain LWB10
|
Biochemical tests |
|
|
Gram reaction |
- |
|
Pigment production |
+ |
|
MacConkey Agar Medium growth |
+ |
|
Growth at 4°C |
- |
|
Growth at 42°C |
- |
|
Catalase test |
+ |
|
Oxidase test |
+ |
|
Nitrite reduction |
- |
|
Arginine dihydrolase |
+ |
|
Assimilation of Glucose |
+ |
|
Mannitol |
+ |
|
Maltose |
+ |
|
Xylose |
+ |
|
Simon's citrate |
++ |
|
Acetamide |
- |
|
DNA |
- |
To provide convincing evidence, we applied other two approaches to identify strain LWB10. Results from MALDI biotypersmart system revealed that LWB10 had a high score value of 2.247 relative to P. mosselii (Fig. 3d). Moreover, we also performed the gas chromatography cellular fatty acid analysis (GC-FA) to identify this strain. Unfortunately, the version of the database is too old that lack of P. mosselii. GC-FA analysis indicated LWB10 was P. putida while P. putida was in the second place of MALDI results and showed a low score value of 1.742 (data not showed). These two procedures together indicated that strain LWB10 is P. mosselii.
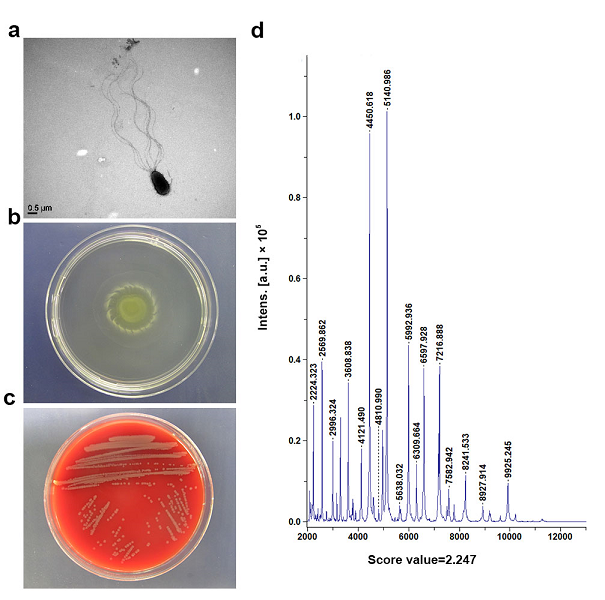
Fig. 3 Morphological, physiological and biochemical characterization of LWB10. (a) Transmission electron microscopy of LWB10, showing polar flagella and rod-shaped cell; (b) Motility activity test of LWB10. (c) Growth on blood agar medium showed a circular and smooth colony; (d) LWB10 profile generated by the high-throughput MALDI Biotypersmart system.
3.4. In vitro antagonism of LWB10 to other A. tumefaciens strains
In order to test the antagonism ability of LWB10 to other A. tumefaciens strains (ACCC19197, CFCC1369, GV3101, LBA4404 and EHA105), we conducted in vitro assays by the inhibition zone test (in 60 mm petri dish). Results showed that LWB10 had diverse inhibition ability to the test strains of A. tumefaciens after 2 days. The diameter of inhibition zones to each strains were 23, 35, 31, 18 and 16 mm (Fig. 4). Strain LWB10 displayed strong antibiotic activity to ACCC19197, CFCC1369 and GV3101. Our results showed that LWB10 has strongly antagonism to other A. tumefaciens strains.
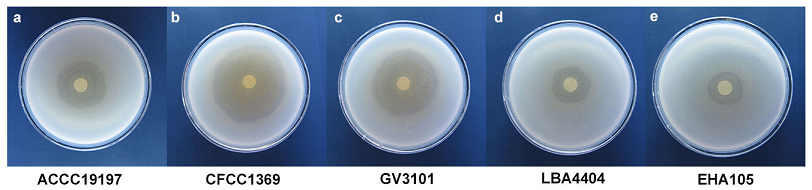
Fig. 4 P. mosselii LWB10 suppresses A. tumefaciens growth in vitro. LWB10 showed significantly growth inhibition of five strains, including (a) ACCC19197, (b) CFCC1369, (c) GV3101, (d) LBA4404 and (e) EHA105. The diameter of inhibition zone was 23, 35, 31, 18 and 16 mm, respectively. Pictures were taken at 2 days after inoculation. Each treatment conducted at least 5 replications.
3.5. Antibiotic activity in the host plant
At 45 dpi, the plants inoculated with CFCC1369 alone showed severe tumor formation in inoculated sites, while in LWB10 added treatments displayed fewer tumor formation in inoculation sites and P. mosselii LWB10 alone showed no symptoms of tumor formation (Fig. 5). The average size of the tumor is 4 × 10 mm in CFCC1369 inoculated stems, and there is few tumors in co-inoculated sets. Symptoms first observed after 30 dpi in tomato main stems of CFCC1369 treated sets, while tumors not format until 40 dpi. Each treatments has at least 15 replications. These results indicated that LWB10 had strong inhibition ability of tumor formation by A. tumefaciens CFCC1369.

Fig. 5 In vivo test of antibiotic activity of LWB10.Main stems of tomato plants were inoculated with CFCC1369 alone, LWB10/CFCC1369 mixture solutions and LWB10 alone. The existence of LWB10 strongly suppressed the tumor formation in tomato stems. Symptoms were taken at 45 dpi. This test was repeated for three times and each time at least 15 replications.
Crown gall disease caused by A. tumefaciens has long been a big threat to agricultural economic. A. tumefaciens was a soil born, gram negative, plant pathogen that exist in the rhizosphere of host plant. After infection, it will produce tumors on infection sites, such as roots and stems. The disease symptoms developed slowly and when the pathogen once established, it will be hard to cure. Increasing evidence has shown that plants recruit protective bacteria to their rhizosphere to enhance microbial activity to suppress pathogens [26-28]. Many researchers have been trying to use biological control method for this disease. Moreover, some other antibiotic strains also found to stop the growth and tumor formation in host plants. As the disease become more and more severe all around the world, we need more new biological control agents to cure this disease.
Among all the 10328 bacterial strains isolated from rhizosphere of cherry tree, only LWB10 strain exhibited strong antibiotic activity to A. tumefaciens. A series of methods, such as biochemical characterization, molecular identification and MALDI-TOF analysis, indicated that LWB10 closely related to P. mosselii. P. mosselii LWB10 was deposited to the China Center for Type Culture Collection (CCTCCas M2019081.
Pseudomonas mosselii regarded as a specie of the P. putida group [29, 30]. It is an environmental species detected in rhizospheric soil and is an overall unusual human opportunistic pathogen [31, 32]. Reports had showed that P. mosselii also has antibacterial ability to other pathogens [33, 34]. Moreover, a novel insecticidal protein, PIP-47Aa, isolated from P. mosselii is toxic to three corn rootworm species. This protein is a novel insecticidal protein for controlling of the corn rootworm pests [35]. P. mosselii has been reported to been characterized in biocontrol against plant disease. Recently, Zhou and colleges generated an engineered P. mosselii strain to express Ralstonia solanacearum ripAA gene, which determines incompatible interactions with tobacco plants to control tobacco bacterial wilt [36]. A gene cluster named c-xtl from P. mosselii BS011 documented to be required for inhibitory activity against the fungus Magnaporthe oryzae [37]. While there are no such reports for the biocontrol of P. mosselii to crown gall formation bacteria Agrobacterium tumefaciens.
Gene deletion experiments demonstrated that the gene cluster named c-xtl from P. mosselii BS011 documented to be required for the inhibitory activity [35]. Engineering of Ralstonia solanacearum ripAA gene to P. mosselii strain is efficient to control tobacco bacterial wilt [36]. A drafted genome sequence of P. mosselii Gil3 predicted for the synthesis of antimicrobial compound xantholysin and a ppyS homologous gene for synthesis of antibiotic pseudopyronines [38]. In this study, we isolated a P. mosselii strain LWB10 that show strong antibiotic activity to A. tumefaciens. And further study is needed to prove out the exact antibiotic agent of P. mosselii LWB10.
The results of the present study provide evidence of the potential antibiotic ability exhibited by P. mosselii LWB10 to control crown gall disease in tomato. It is also the first report on the antibiotic ability of P. mosselii LWB10 to A. tumefaciens. Future research should focus on extraction assays to prove out the exact antibiotic agent of P. mosselii LWB10. P. mosselii LWB10 provided a new strain for the biocontrol of plant crown gall disease.
This work is supported by the Natural Science Foundation of Ningbo, China (2018A610215), Major agricultural project of Ningbo (2016C11011), the Key Projects of Ningbo City College of Vocational Technology (ZZX18122) and the Key Projects of Ningbo City College of Vocational Technology (ZZX16047).
De Cleene M, and Deley J. (1976) Host range of crown gall. Bot Rev 42:389-466
View ArticleConner AJ, and Dommisse EM (1992) Monocotyledonous plants as hosts for Agrobacterium. Int J Plant Sci 153: 550-555
View ArticleDe Cleene M. (1979) Crown gall: economic importance and control. Zentralblatt fur Bakteriologie, Parasitenkunde, Infektionskrankheiten und Hygiene. Zweite naturwissenschaftliche Abteilung: Mikrobiologie der Landwirtschaft der Technologie und des Umweltschutzes 134: 551-554 80080-4
View ArticleEscobar MA, Civerolo EL, Polito VS, Pinney KA, Dandekar AM (2003) Characterization of oncogene-silenced transgenic plants: implications for Agrobacterium biology and post-transcriptional gene silencing. Mol Plant Pathol 4: 57-65 PMid:20569363
View Article PubMed/NCBIGelvin SB (1990) Crown gall disease and hairy root disease - a sledgehammer and a tackhammer. Plant Physiol 92: 281-285 PMid:16667272
View Article PubMed/NCBIRhouma A, Bouri M, Boubaker A, Nesme X (2008) Potential effect of rhizobacteria in the management of crown gall disease caused by Agrobacterium tumefaciens biovar 1. J Plant Pathol 90: 517-526
Hammami I, Rhouma A, Jaouadi B, Rebai A, Nesme X (2009) Optimization and biochemical characterization of a bacteriocin from a newly isolated Bacillus subtilis strain 14B for biocontrol of Agrobacterium spp. strains. Lett Appl Microbiol 48: 253-260 PMid:19196444
View Article PubMed/NCBIHammami I, Jaouadi B, Ben Bacha A, Rebai A, Bejar S, Nesme X, Rhouma A (2012) Bacillus subtilis bacteriocin Bac 14B with a broad inhibitory spectrum: Purification, amino acid sequence analysis, and physicochemical characterization. Biotechnol Bioproc E 17: 41-49
View ArticleFilo A, Sabbatini P, Sundin GW, Zabadal TJ, Safir GR, Cousins PS (2013) Grapevine crown gall suppression using biological control and genetic engineering: A Review of Recent Research. Am J Enol Viticult 64: 1-14
View ArticleBen Abdallah D, Frikha-Gargouri O, Tounsi S (2015) Bacillus amyloliquefaciens strain 32a as a source of lipopeptides for biocontrol of Agrobacterium tumefaciens strains. J Appl Microbiol 119: 196-207 PMid:25764969
View Article PubMed/NCBIKerr A (1972) Biological-control of crown gall - seed inoculation. J Appl Bacteriol 35: 493-496
View ArticleFarkas E, Haas JH (1985) Biological-control of crown gall in rose nursery stock. Phytoparasitica 13: 121-127
View ArticleMoore LW, Tingey DT (1976) Effect of temperature, plant age, and infection site on severity of crown gall disease in radish. Phytopathol 66: 1328-1333
View ArticleFrikha-Gargouri O, Ben Abdallah D, Ghorbel I, Charfeddine I, Jlaiel L, Triki MA, Tounsi S (2017) Lipopeptides from a novel Bacillus methylotrophicus 39b strain suppress Agrobacterium crown gall tumours on tomato plants. Pest Manag Sci 73: 568-574 PMid:27252109
View Article PubMed/NCBIHolloway BW, Krishnapillai V, Morgan AF (1979) Chromosomal genetics of Pseudomonas. Microbiol Rev 43: 73-102 PMid:111024
View Article PubMed/NCBISilby MW, Winstanley C, Godfrey SAC, Levy SB, Jackson RW (2011) Pseudomonas genomes: diverse and adaptable. Fems Microbiol Rev 35: 652-680 PMid:21361996
View Article PubMed/NCBIPoblete-Castro I, Becker J, Dohnt K, dos Santos VM, Wittmann C (2012) Industrial biotechnology of Pseudomonas putida and related species. Appl Microbiol Biot 93: 2279-2290 PMid:22350258
View Article PubMed/NCBIGhequire MGK, De Mot R (2014) Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol Rev 38: 523-568 PMid:24923764
View Article PubMed/NCBINikel PI, Martinez-Garcia E, de Lorenzo V (2014) Biotechnological domestication of Pseudomonads using synthetic biology. Nat Rev Microbiol 12: 368-379 PMid:24736795
View Article PubMed/NCBIGross H, Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26: 1408-1446 PMid:19844639
View Article PubMed/NCBIXu XQ, Pan SQ (2000) An Agrobacterium catalase is a virulence factor involved in tumorigenesis. Mol Microbiol 35: 407-414 PMid:10652101
View Article PubMed/NCBILi W, Cao J-Y, Xu Y-P, Cai X-Z (2017) Artificial Agrobacterium tumefaciens strains exhibit diverse mechanisms to repress Xanthomonas oryzae pv. oryzae-induced hypersensitive response and non-host resistance in Nicotiana benthamiana. Mol Plant Pathol 18: 489-502 PMid:27061769
View Article PubMed/NCBIMurray RGE, Doetsch RN, Robinow CF (1994) Determinative and cytological light microscopy. Methods Gen Mol Bacteriol 21-41
Turvey ME, Weiland F, Meneses J, Sterenberg N, Hoffmann P (2016) Identification of beer spoilage microorganisms using the MALDI Biotyper platform. Appl Microbiol Biotechnol 100: 2761-2773 PMid:26857464
View Article PubMed/NCBIKumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol 33: 1870-1874 PMid:27004904
View Article PubMed/NCBIBerendsen RL, Pieterse CMJ, Bakker PAHM (2012) The rhizosphere microbiome and plant health. Trends in Plant Sci 17: 478-486 PMid:22564542
View Article PubMed/NCBIMendes R, Garbeva P, Raaijmakers JM (2013) The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634-663 PMid:23790204
View Article PubMed/NCBIChen Y, Wang J, Yang N, Wen Z, Sun X, Chai Y, Ma Z (2018) Wheat microbiome bacteria can reduce virulence of a plant pathogenic fungus by altering histone acetylation. Nat Commun 9 PMid:30143616
View Article PubMed/NCBIDabboussi F, Hamze M, Singer E, Geoffroy V, Meyer JM, Izard D (2002) Pseudomonas mosselii sp nov., a novel species isolated from clinical specimens. Int J Syst Evol Microbiol 52: 363-376 PMid:11931144
View Article PubMed/NCBIMulet M, Lalucat J, Garcia-Valdes E (2010) DNA sequence-based analysis of the Pseudomonas species. Environ Microbiol 12: 1513-1530 PMid:20192968
View Article PubMed/NCBILeneveu-Jenvrin C, Madi A, Bouffartigues E, Biaggini K, Connil N (2013) Cytotoxicity and inflammatory potential of two Pseudomonas mosselii strains isolated from clinical samples of hospitalized patients. BMC Microbiol 13, 123. PMid:23718251
View Article PubMed/NCBIGiani T, Marchese A, Coppo E, Kroumova V, Rossolini GM (2012) VIM-1-producing Pseudomonas mosselii isolates in Italy, predating known VIM-producing index strains. Antimicrob Agents Chemother. 56(4):2216-2217. PMid:22290983
View Article PubMed/NCBIKhmel IA, Sorokina TA, Lemanova NB, Lipasova VA, Metlitski OZ, Burdeinaya TV, Chernin LS (1998) Biological control of crown gall in grapevine and raspberry by two Pseudomonas spp. with a wide spectrum of antagonistic activity. Biocontrol Sci Technol 8: 45-57
View ArticleDandurishvili N, Toklikishvili N, Ovadis M, Eliashvili P, Giorgobiani N, Keshelava R, Tediashvili M, Vainstein A, Khmel I, Szegedi E, Chernin L (2011) Broad-range antagonistic rhizobacteria Pseudomonas fluorescens and Serratia plymuthica suppress Agrobacterium crown gall tumours on tomato plants. J Appl Microbiol 110: 341-352 PMid:21091861
View Article PubMed/NCBIWei J-Z, O'Rear J, Schellenberger U, Rosen BA, Park Y-J, McDonald MJ, Zhu G, Xie W, Kassa A, Procyk L, Ortega CP, Zhao J-Z, Yalpani N, Crane VC, Diehn SH, Sandahl GA, Nelson ME, Lu AL, Wu G, Liu L (2018) A selective insecticidal protein from Pseudomonas mosselii for corn rootworm control. Plant Biotechnol J 16: 649-659 PMid:28796437
View Article PubMed/NCBIZhou T, Chen ST, Fan XJ, Hu X, Zou, HS (2019) An improved control efficacy against tobacco bacterial wilt by an engineered Pseudomonas mosselii expressing the ripAA gene from phytopathogenic Ralstonia solanacearum. BioRxiv
View ArticleWu L, Xiao W, Chen G, Song D, Khaskheli MA, Li P, Zhang S, Feng G (2018) Identification of Pseudomonas mosselii BS011 gene clusters required for suppression of Rice Blast Fungus Magnaporthe oryzae. J Biotechnol 282: 1-9 PMid:29704539
View Article PubMed/NCBIZhang D, Xu D-H, Qiu J, Rasmussen-Ivey CR, Liles MR, Beck BH (2016) Draft Genome Sequence of Pseudomonas mosselii Gil3, Isolated from Catfish and Antagonistic against Hypervirulent Aeromonas hydrophila. Genome Announc 4 PMid:27856595
View Article PubMed/NCBI