LIANG Zong-ying
Email: liangzy0318@163.com
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 1
Page No: 220-227
LIANG Zong-ying
Email: liangzy0318@163.com
LIANG Zong-yingΔ, HUANG Jing-tao
Department of Thoracic Surgery, The Affiliated Hospital of Chengde Medical University, Heibei Chengde 067000,China
LIANG Zong-ying, HUANG Jing-tao, NU9056 targets KAT5 to regulate the Proliferation, migration and invasion of esophageal cancer cells via ABCE1 acetylation(2022)Journal of Cellular and Molecular Physiology 4(1):220-227
Objective: To explore the mechanism by which NU9056 affects the epithelial mesenchymal transformation (EMT), proliferation, migration, and invasion by regulating ABCE1 protein acetylation through KAT5.
Methods: TE-1 cells were cultured in vitro, NU9056 concentration by MTT, KAT5 mRN by qRT-PCR, ABCE1 acetylation by immunoprecipitation, KAT5 and EMT-related proteins by Western blot, MTT, migration, and invasion by Transwell.
Results: The optimal administration concentration of NU9056 was 1.0 µmol/L via the MTT. In the NU9056 group, KAT5 mRNA and protein expression decreased, and ABCE1 acetylation level decreased (P<0.05); in the NU9056 group, EMT marker protein E-cadherin was downregulated, while N-cadherin and Slug proteins were downregulated (P<0.05). TE-1 cell survival, migration, and invasion were significantly decreased in the NU9056 group (P<0.05).
Conclusion: NU9056 may reduce the ABCE1 protein acetylation levels by downregulating KAT5 expression, and subsequently inhibit the EMT, survival, migration, and invasion capacity of esophageal cancer cells.
Key words: Acetyltransferase inhibitor NU9056 KAT5 ABCE1, modified by acetylation
Esophageal cancer is one of the most aggressive gastrointestinal cancers, greatly endangering the health of people around the world[1]. Although treatments including surgical resection, chemotherapy and radiotherapy have improved the prognosis of patients with esophageal cancer somewhat, their overall survival rate and quality of life are still poor [2].The ATP binding box transporter E1 (ATP combined box transporter E1, ABCE1) is an oncogene, whose expressed ABCE1 protein is highly expressed in many tumors, and it is closely related to the tumor proliferation, metastasis and prognosis[3]. ABCE1 was found to be highly expressed in oesophageal cancer tissues, and it is closely related to the proliferation, apoptosis, and invasion and metastasis of oesophageal cancer[4]. However, the specific mechanism by which ABCE1 promotes the occurrence and development of oesophageal cancer remains unclear. Previous studies showed that ABCE1 was highly expressed and acetylated in esophageal cancer tissues and metastatic lymph nodes, and its high acetylation level was associated with esophageal cancer stage, tissue differentiation and lymph node metastasis[5]. Acetylation modifications in epigenetics are one of the major post-translational modifications of proteins, and cross-regulation between acetylation modifications and other modifications is essential for the transcriptional control of genes[6]. The acetylation modification of proteins requires the catalysis of acetyltransferases, and the protein function after the acetylation modification changes and strengthens, participating in the tumor initiation and development[7]. KAT5 is an important member of the acetyltransferase family, which can catalyze the acetylation of various histones and non-histones, and then participate in the occurrence and development of tumors[8]. Protein acetylation modification requires the participation of acetyltransferase. KAT5 as a classical acetyltransferase can participate in tumor development and metastasis by regulating protein acetylation modification, but its role in related protein acetylation modification in esophageal cancer has not been reported. In this study, we observed the effect of acetyltransferase inhibitor NU9056 on the level of acetyltransferase KAT5 and ABCE1 acetylation in esophageal cancer cells, and explored the possible intrinsic connection between KAT5 and ABCE1 acetylation and the possible mechanism of action in the development and development of esophageal cancer.
2.1 Materials and instruments
The human oesophageal cancer cell line TE-1 was purchased from Biotechnology Development Co.Ltd. (Shanghai,China) and maintained in a 37℃ incubator containing 5% CO2. NU9056 was purchased from Biotechnology Development Co.Ltd. (Shanghai, China); RPMI 1640 and PBS were purchased from Bioindustry (Beijing, China); FBS was purchased from Biotech Corporation (Shanghai, China); ABCE1 monoclonal antibody and Acetylated-Lysine antibodies were purchased from Biotech (Abcam, USA) ; β-actin polyclonal antibody was purchased from Beijing Zhongshan Jinqiao Biotechnology Company; A/G agarose beads were purchased from Biotech Corporation (CST, China); CO2 incubator and Doska FC microplate reader were purchased from Heat (USA); Faith Caliper was purchased from BD Corporation (USA).
2.2 Experimental method
2.2.1 Cell culture
Esophageal cancer TE-1 cells were cultured in RPMI 1640 medium containing 10% FBS in a 37℃, 5% CO2 incubator, pass1:3 when density reached around 80%, and vigorously growing log TE-1 cells were used for subsequent experiments.
2.2.2 Relative mRNA expression was measured by RT-qPCR
RNA was extracted and reverse transcribed to complementary DNA using the RNA reverse transcription kit. The expression of mRNA was detected by the SYBR Green qPCR Master fluorescence quantification kit; reaction conditions for PCR: 94℃ predenaturation for 4min, 94℃ denaturation for 30s; 55℃ annealing for 30s; and 72℃ extension for 45s (30 cycles). The dissolution curve refers to the instrument automatic procedure. Relative expression was calculated using U6 and GAPDH, and the internal parameters was calculated by 2- ΔΔ Ct.
2.2.3 Immunoprecipitation experiments to detect protein acetylation
Total cellular protein was extracted, and the concentration was determined. After adding 5 µl of target protein pure antibody and 5ml of A/G agarose beads, 2 lysis buffer was supplemented to a total volume of 450 µl. After centrifugation, the supernatant was removed in 400 µl into centrifuge tubes for 4℃ and 15 r/min for 12h. It was centrifuged at 4℃ and the supernatant was discarded. A/G agarose beads were washed in 500 µl with 1× lysis buffer. The mixture was centrifuged for 3 min and the supernatant was discarded. Repeat the wash 3 times; discard the supernatant after the last wash.1×Lysis buffer 35 µl and an equal volume of 2×SDS loading buffer were mixed, boiled and centrifuged; the next walk Western blot experiment.
2.2.4 Protein expression was measured by the Western blot experiments
Total protein was extracted and determined by protein concentration; it was separated by gel electrophoresis and transferred to the PVDF membrane by the semi-dry transmembrane method. After blocking the 5% skim milk powder for 1h at room temperature, the membrane was incubated in a diluted primary antibody for 40 min. After rinsing, the membrane was incubated in a diluted secondary antibody for 30 min. After rinsing, the luminescence reaction was performed in the dark chamber and exposed into slices. The integrated optical density IOD values of the protein bands were analyzed by Image proplus software to reflect the expression amount of the target proteins in each group as the ratio of the IOD value of the target protein and the IOD value of the internal parameters.
2.2.5 Cell viability was determined by the MTT assay
Single-cell suspension was followed at 8103 cells per well were seeded in 96-well plates. Cultures were routinely grown until the cell density reached 80% per well for transfection. After 12, 24, 48 and 72 hours, a plate was removed at each time point, and 5 mg/ml of MTT solution of 30 µ1 was added to each well, and the conventional incubation was continued for 4 hours. Centrifugation, abandon the upper clear. Dimethylsulfoxide (DMSO) was added at 150 µ1 to each well and shaken with an 80 r/min horizontal shaker for 5-10 minutes. The absorbance value (OD value) of 570nm per well was measured by a microplate meter.
2.2.6 Scratch repair assay was used to detect cell migration ability
Cells from each group were collected and paired with a single-cell suspension; 8,103 cells per well were seeded into 96-well plates.100 µ1 per well in three groups. Conventional culture, after the bottom of the sterile pipette head vertical plate scratches. 2 ml of serum-free medium was added to each well for a further 48 h of culture and the width of the scratches was measured under an inverted microscope. Cell mobility = (scratch width / initial scratch width) 100%.
2.2.7 Transwell chamber assay was used to detect cell invasion ability
Cells covered at the pore bottom after transfection were taken and digested as a single cell suspension with trypsin of 0.25% without EDTA, and cells were harvested and counted after collection. The Matrigel glue melts into a liquid state early. The 50 mg/L Matrigel 1:8 dilution solution was coated with a polycarbonate microporous filter membrane at 8 µm aperture at the bottom of the Transwell chamber and polymerized into glue at 37℃ overnight. The chamber was UV-sterilized for 2h, and the residual Matrigel gel solution was removed and moistened with serum-free F12 medium for 1h at 37℃. The Transwell chamber was placed sequentially into a 24-well plate, sterile forceps were removed, and 600 µ1 of 15% serum F12 medium was added outdoors. In the small chamber, 200 µ1 of cell suspension (cell number: 2104) was added, the culture medium was F12 medium without serum, and 6 samples were repeated for each group of cells. 24-well plates were routinely cultured in CO2 incubation tanks for 48h. The chamber was removed, rinsed by PBS, and the inner layer of the microporous membrane erased. Crystal violet staining after paraformaldehyde fixation. Photo counting under the microscope.
2.3 Statistical Methods
SPSS21.0 statistical analysis software was used. Measurement data were expressed as mean±standard deviation. One way ANOVA was used for statistical analysis. And a value of P < 0.05 was considered statistically significant.
3.1. NU9056 concentration-dependent inhibited the viability of TE-1 cells in esophageal cancer
The results of MTT experiment showed that NU9056 significantly inhibited the proliferation vitality of TE-1 cells in esophageal cancer (P<0.05), and the semi-inhibitory concentration was IC50=1.073 µmol/L. Determine 1.0 µmol/L as the optimal dosing concentration of NU9056. ( Fig. 1)
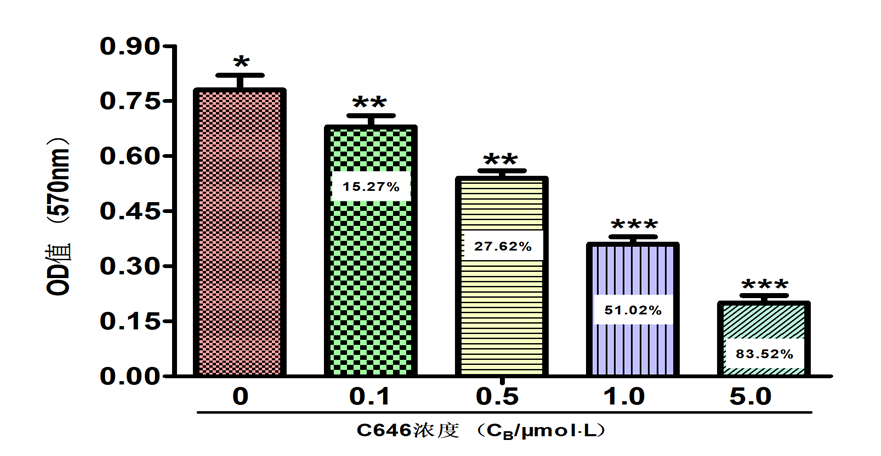
Fig 1. The effect of NU9056 on cell viability of TE-1 in esophageal cancer was determined by M T T assay
3.2. NU9056 inhibits KAT5 mRNA and protein expression
The RT-qPCR results showed that the relative expression of KAT5 mRNA in the NU9056 group was (0.19±0.05) and the DMSO group was (1.18±0.04), and the NU9056 group was significantly reduced and statistically significant (P<0.05). The Western blot experiment results showed that the KAT5 protein expression was significantly reduced in the NU9056 group, and the two groups were statistically significant (P<0.05). ( Fig. 2)
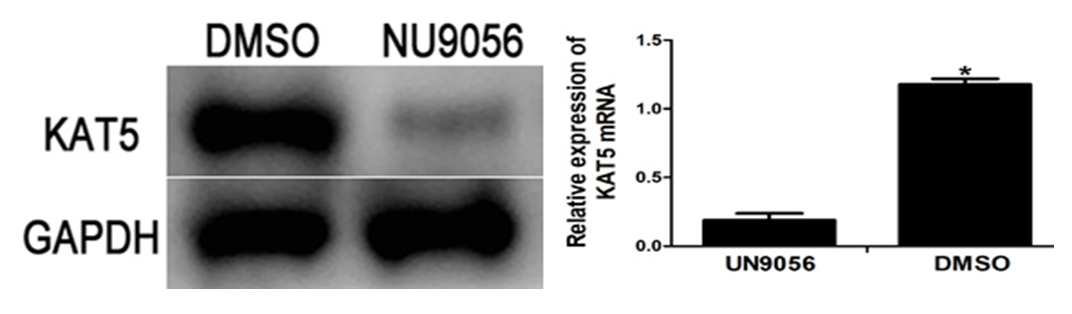
Fig 2. Effect of NU9056 on KAT5 mRNA and protein expression in esophageal cancer cells
3.3. NU9056 reduces the protein acetylation level of ABCE1
Co-immunoprecipitation and Western Blot assays showed decreased KAT5 protein expression and decreased ABCE1 protein acetylation levels in the NU9056 group. The difference was significant from the DMSO group (P<0.05).( Fig. 3)
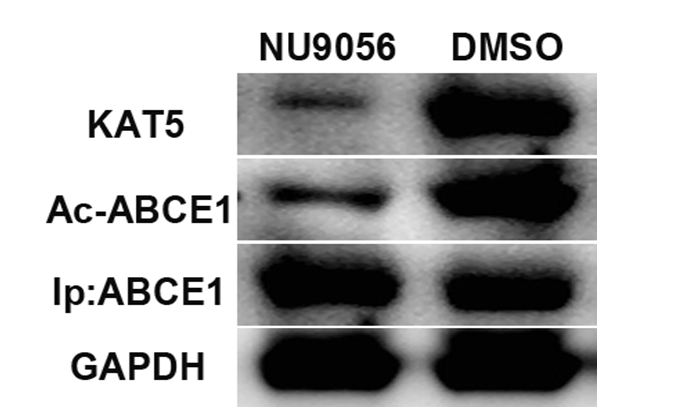
Fig 3. Effect of NU9056 on the acetylation levels of ABCE1 protein in oesophageal cancer cells
3.4. NU9056 inhibits EMT in esophageal cancer TE-1 cells
The Western blot results showed that NU9056 could significantly upregulate the expression of the epithelial cell marker protein E-cadherin, N-cadherin and the mesenchymal cell marker protein and EMT-related transcription regulator Slug in TE-1 cells compared with the DMSO group, with statistically significant differences between the two groups.( Fig. 4)
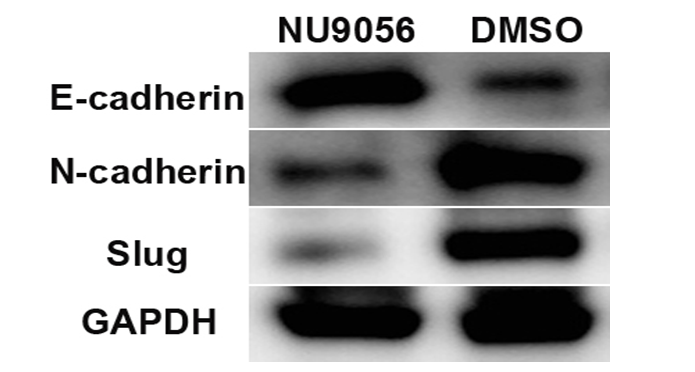
Fig 4. NU9056 effect on the expression of EMT-associated protein in esophageal cancer cells
3.5 NU9056 inhibits the viability of TE-1 cells in oesophageal cancer
The results of the MTT experiments showed that the absorbance values of TE-1 cells were significantly reduced in the NU9056 group, which inhibited the viability of TE-1 cells. Significant difference between the two groups was observed (P<0.05).( Fig. 5)
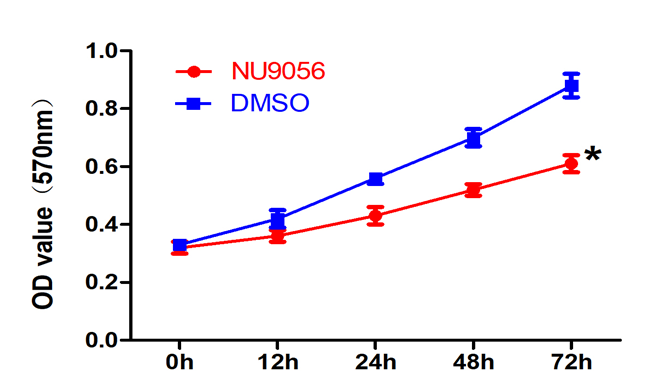
Fig 5. Effect of NU9056 on the viability of TE-1 cells in esophageal cancer
3.6. NU9056 inhibits the migratory ability of TE-1 cells in oesophageal cancer
The results of the cell scratch healing assay showed that the scratch healing was slow and the distance was slightly shortened in the NU9056 group, which inhibited the migration ability of TE-1 cells. Significant difference between the two groups was observed (P<0.05).( Fig. 6)
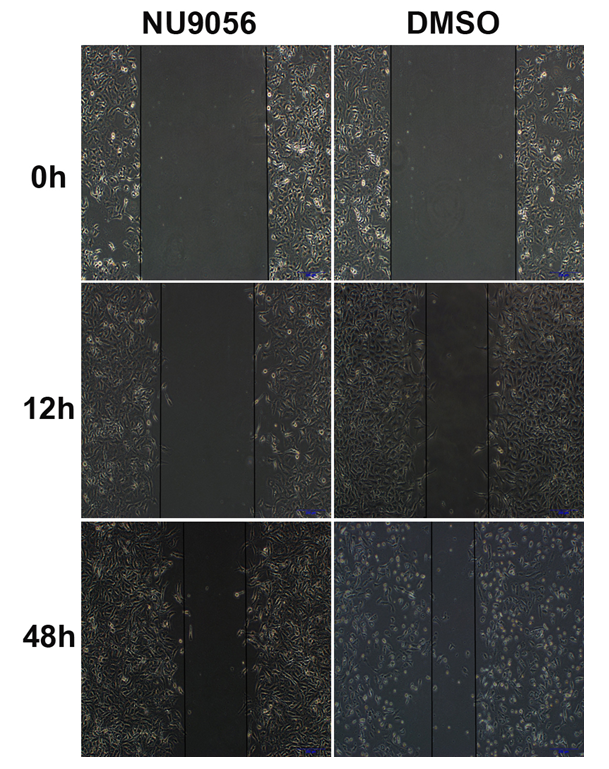
Fig 6. Effect of NU9056 on the migratory capacity of TE-1 cells in esophageal cancer
3.7. NU9056 inhibits TE-1 invasion ability in oesophageal cancer
The results of Transwell chamber experiment showed that the number of membrane cells was significantly reduced in NU9056 group, which inhibited the invasive ability of TE-1 cells. Significant difference between the two groups was observed (P<0.05).( Fig. 7)
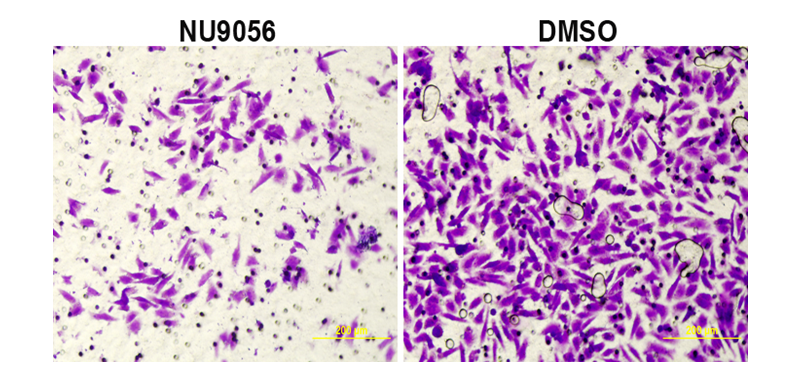
Fig 7. Effect of NU9056 on the invasive capacity of TE-1 cells in esophageal carcinoma
The overall prognosis of oesophageal cancer is poor, with a 5-year survival rate of only 20% to 35%, and despite advances in surgical and chemoradiotherapy regimen optimization, the overall benefit is very little for [9]. The high invasive and metastatic nature of tumors is the main cause of the high mortality of patients, and both the oncogene activation and the inactivation of tumor suppressor genes are the key factors leading to the failure of tumors in clinical treatment[10]. Therefore, to effectively inhibit the survival ability of cancer cells and one of the important methods to improve the long-term survival rate is to prevent the invasion and metastasis of esophageal cancer.
ABCE1 is highly expressed in a variety of malignant tumors and is considered as an oncogene, which is closely related to the occurrence and development of a variety of malignant tumors (10). Recent studies have found that the expression of ABCE1 in lung cancer tissues is higher than that in normal lung tissues, which is closely related to the proliferation, invasion and metastasis of NSCLC [11,12]. ABCE1 expression is significantly increased in gastric cancer tissues, which is involved in the proliferation, migration and apoptosis of gastric cancer cells [13]. Studies have shown that protein acetylation can lead to changes in protein function, induce tumors, and promote tumor development. At present, there is no relevant report on the acetylation of ABCE1 protein in esophageal cancer and the effect of acetylation on esophageal cancer. Previous studies of our group have shown that ABCE1 protein is highly expressed in esophageal cancer tissues, while it is low expressed in normal esophageal mucosa. Moreover, ABCE1 is also acetylated in esophageal cancer tissues, and its acetylation level is significantly correlated with TNM staging, tumor differentiation and lymph node metastasis of esophageal cancer[14]. This indicates that ABCE1 is an acetylated protein, and its acetylation modification may be involved in the proliferation, invasion and metastasis of esophageal cancer.
The acetylation of proteins requires the catalysis of acetyltransferase, and the function of acetylated proteins will be changed and strengthened, which is involved in the occurrence and development of tumors (15). Acetyltransferase KAT5 is an important member of the acetyltransferase family, which can catalyze the acetylation of various histones and non-histones. Previous experimental studies have confirmed that KAT5 is highly expressed in esophageal cancer TE-1 cells, and its high expression is synchronized with the high acetylation state of ABCE1 protein, which indicates that KAT5 and ABCE are also involved in the occurrence and development of esophageal cancer. However, whether KAT5, as an acetyltransferase in esophageal cancer, can catalyze the acetylation of ABCE1 to regulate the occurrence and development of esophageal cancer needs to be further verified. Theoretically, inhibition of acetyltransferase expression can change the acetylation status of ABCE1 protein in esophageal cancer tissue, and then change the biological behavior of esophageal cancer. In the mechanism of silencing or downregulating acetyltransferase to promote the deacetylation of related proteins, silencing KAT5 may play a higher role in deacetylation. However, whether KAT5, as an acetyltransferase, is involved in the acetylation of ABCE1 in esophageal cancer remains unclear.
NU9056, as an acetyltransferase inhibitor, reduced the levels of acetylated histones H4K16, H3K14 and H4K8 in LNCaP cells, which are targets of KAT5-mediated acetylation. In this study, esophageal cancer TE-1 cells were treated with NU9056. MTT assay confirmed that NU9056 could inhibit the proliferation of esophageal cancer TE-1 cells in a concentration-dependent manner. Acetyltransferase inhibitors can inhibit a variety of acetyltransferase activities in vivo, but they usually have a targeted mainly inhibit the activity and expression of one acetyltransferase. The expression of acetyltransferase KAT5 in TE-1 cells treated with NU9056 was detected by RT-PCR and Western Blot. The results showed that NU9056 could significantly inhibit the expression of KAT5 mRNA and protein. This indicates that NU9056 can inhibit the activity and expression of KAT5 and is a specific inhibitor of KAT5. Further detection of ABCE1 acetylation level showed that ABCE1 acetylation in esophageal cancer cells was significantly reduced after down-regulation of KAT5 expression, suggesting that KAT5, as an acetyl transferase, is involved in the acetylation of ABCE1, which may be an important upstream molecule regulating ABCE1 and then changing the biological function of esophageal cancer.
Epithelial mesenchymal transformation (EMT) is an important mechanism to promote tumor invasion and metastasis. Tumor cells gradually lose cell polarity and adhesion, and then gain high migration ability of mesenchymal cells, thus accelerating tumor invasion and metastasis. It is an early marker of invasion and metastasis of various malignant tumors [16]. EMT plays a key role in the process of tumor invasion and migration, mainly including E-cadherin, N-cadherin and Slug protein expression, which is also an important marker for predicting tumor invasion and metastasis [17]. The expression and function of EMT marker proteins and related transcriptional regulators such as Slug are regulated by post-translational modifications such as acetylation, phosphorylation and ubiquitination, which subsequently mediate the occurrence and development of tumors [18]. This study confirmed that the expression of E-cadherin was up-regulated in esophageal cancer cells after NU9056 intervention, while the expression of N-cadherin and Slug proteins was down-regulated. In addition, NU9056 inhibited the survival, migration and invasion of esophageal cancer cells by cell functional tests. These results indicate that NU9056 inhibits the activity of acetyltransferase KAT5 and deacetylates ABCE1 protein, which regulates the expression of EMT marker proteins and related transcription factors, and then inhibits the survival, invasion and migration of esophageal cancer TE-1 cells.
In conclusion, NU9056, an acetyltransferase inhibitor, can regulate the acetylation of ABCE1 by down-regulating acetyltransferase KAT5, and further inhibit the proliferation, invasion and migration of esophageal cancer by regulating key factors in the process of EMT, which provides a new method for the clinical treatment of esophageal cancer.
Funding
Hebei Province Medical Science Research Key Project Plan (Project No.: 20181159)
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this paper.
Watanabe M, Otake R, Kozuki R, et al. Recent progress in multidisciplinary treatment for patients with esophageal cancer. Surg Today[J]. 2020 Jan; 50 (1) : 12 to 20. PMid:31535225
View Article PubMed/NCBIDudash M, Slipak S,Dove JT,et al.Lymph Node Harvest as a measure of quality and effect on overall survival in the cancer: A national cancer database assessment[J]. Am Surg,2019,85(2):201-205. PMid:30819299
View Article PubMed/NCBILiang Z, Yu Q, Ji H, Tian D. Tip60-siRNA regulates ABCE1 acetylation to suppress lung cancer growth via activation of the apoptotic signaling pathway[J]. Exp Ther Med. 2019 Apr; 17 (4) : 3195-3202.
View ArticleLiang ZY, Sun GR, Zhao BS, et al. Expression and clinical significance of adenosine triphosphate binding cassette transporter E1 and P300/CBP related factors in esophageal squamous cell carcinoma [J]. Shaanxi medical journal,2020,49(10):1356-1358.(in Chinese)
Liang ZY, Hou JS, Zhang L, et al. Correlation between Survivin acetylation and PCAF expression in esophageal carcinoma [J]. Journal of clinical research in medicine,2019,36(8):1475-477.(in Chinese)
Narita T, Weinert BT, Choudhary C. Functions and mechanisms of non-histone protein acetylation. Nat Rev Mol Cell Biol. 2019 Mar; 20 (3) : 156-174. PMid:30467427
View Article PubMed/NCBIDing JL, Zhang ZP, Liu J, et al. Research progress of multi-target antitumor drugs based on histone deacetylase [J]. Chinese journal of new drugs,2021,30(3):236-245. (in Chinese)
Zhang H, Zhang SS, Zhou PK. Research progress of acetyltransferase Tip60(KAT5) function [J]. Progress in biochemistry and biophysics,2015,42(1):25-31. (in Chinese)
Kelly RJ. Emerging Multimodality Approaches to Treat Localized Esophageal Cancer[J]. J Natl Compr Canc Netw. 2019 Aug 1; 17 (8) : 1009-1014. PMid:31390584
View Article PubMed/NCBITaieb J,Andre T,Auclin E.Refining adjuvant therapy for nonmetastatic colon cancer,new standards and perspectives[J]. Cancer Treat Rev, 2019,75 (28) : 1-11. PMid:30849607
View Article PubMed/NCBIWu Zhuo, Field, Yu Qian, et al. Expression and clinical significance of CCL7 and ABCE1 in NSCLC lung cancer tissues [J]. Modern Oncology Medicine, 2021, 29 (16) : 2837-2840.
Zhou Di, Field, Yang Xueying. Regulation of miR-203 on ABCE1 during the progression of non-SCLC [J]. Journal of China Medical University, 2021, 50 (6) : 548-552.
Ding D, Ao X, Li M, et al. FOXO3a-dependent Parkin regulates the development of gastric cancer by targeting ATP-binding cassette transporter E1 [J]. J Cell Physiol, 2021236 (4) : 2740-2755. PMid:32914432
View Article PubMed/NCBILiang ZY, Yang Y, Sun GR, et al. Relationship between acetyltransferase KAT3b and ABCE1 acetylation in esophageal cancer [J]. Chin J Cancer, 222,32(04):316-323.(in Chinese)
Ding JL, Zhang ZP, Liu J, et al. Advances in multi-target antitumor drugs based on histone deacetylase design [J]. The Chinese Journal of New Drugs, 2021, 30 (3) : 236-245.
Franke FC, Muller J, Abal M, et al. The tumor suppressor SASH1 interacts with the signal adaptor CRKL to inhibit epithelial -mesenchymal transition Metastasis in colorectal cancer [J]. Cell Mol Gastroenterol Hepatol,2019,7(1):33-53. PMid:30480076
View Article PubMed/NCBIZhou LL, Ge XG, Cai MH, et al. Correlation between ETS-1 and expression of E-cadherin, N-cadherin and Vimentin in non-small cell lung cancer [J]. Cancer Progression,2015,(5):504-511.(in Chinese)
Meng J, Liu XL, Che L, et al. Effects of trcomycin A on epithelial mesenchymal transition, invasion and metastasis of colon cancer HCT116 cells by regulating Ku70 acetylation level [J]. Journal of military medicine,2019,43(02):140-145.(in Chinese)
