Qihan Li
Tel: 86-871-68335905. Fax: 86-871-68334483, Email: liqihan@imbcams.com.cn
microRNA expression profiling in human diploid cells infected with enterovirus 71 or coxsackievirus A16
Corresponding Author
Affiliation
Yao Gao1,2,3#, Ying Zhang1,#, Jianbin Wang1,3, Yanchun Che1, Jingjing Wang1, Xingli Xu1, You Gao1, Yun Liao1, Min Feng1, Shengtao Fan1, Zengfu Yang1, Xinqiang Deng1, Qihan Li1*
- Institute of Medical Biology, Chinese Academy of Medicine Sciences and Peking Union Medical College, Yunnan Key Laboratory of Vaccine Research and Development on Severe Infectious Diseases, Kunming, 650118, China.
- Chuxiong Medical College of Yunnan Province, Chuxiong, 675005, China
- Kunming Medical University, Kunming, 650500, China
Article Reviewed By:
Mingqi Luo(mingqiluo@163.com)
Jos%C3%A9 I N%C3%BA%C3%B1ez(joseignacio.nunez@irta.cat)
Xiaojun Hu(hux4@mail.nih.gov)
Jing Wu(wujing36@163.com)
Citation
Ying Zhang, microRNA expression profiling in human diploid cells infected with enterovirus 71 or coxsackievirus A16(2018)SDRP Journal of Cellular and Molecular Physiology 2(2)p:129-143
Abstract
Background: Enterovirus 71 (EV71) and coxsackievirus A16 are the main causative agents of hand, foot and mouth disease (HFMD), but their pathogenicity has been shown to exhibit wide diversity.
Methods: To illuminate the diverse pathogenicity of EV71 and CA16, microRNA (miRNA) profiles of human diploid cells infected with these two viruses were analyzed using an eukaryotic miRNA expression array.
Results: Unlike EV71, directly stimulating an effective immune response, CA16 infection failed to effectively activate cytokines or the B-/T-cell immune response by repressing the expression of miRNAs. Similarly, the cytokines and antiviral-specific immune responses induced by CA16 infection were lower than those induced by EV71 in rhesus macaques. Individual miRNAs induced by EV71 and CA16 infections modulate the immune-associated target genes and lead to a distinct immune response.
Conclusion: Our results provide experimental evidence focused on the miRNAs induced during EV71 and CA16 infection of human fibroblasts, which suggests some meaningful clues for explanation the wide diversity of immune responses induced by the two viruses.
Keywords: microRNA, human diploid cells, enterovirus 71, coxsackievirus A16
Introduction
Millions of cases of hand, foot and mouth disease (HFMD) and several hundred HFMD-associated deaths have been reported each year in China since 2008, when there was a large outbreak of HFMD in children in Fuyang, Anhui Province [1]. Etiological studies of these cases have demonstrated that HFMD could be frequently caused by pathogens, including enterovirus71 (EV71), coxsackievirus A16 virus (CA16) and echoviruses [2, 3], which are members of the genus Enterovirus. Interestingly, epidemiological data on HFMD have demonstrated that the pathogenicity of the different viruses, which all belong to the family Picornaviridae, is different in the context of recurrent infections. EV71 generally leads to severe cases associated with the nervous system [4-6] and triggers a lower re-infection rate in patients with a history of previous EV71 infection [7]. Conversely, some studies have suggested the possible development of recurrent CA16 infections [7, 8]. However, the few details on the mechanism of CA16 infection [9-12] and the limited knowledge regarding the pathogenicity of HFMD [13, 14] make the potential reason for the infections unclear.
The immunologic antiviral mechanisms induced by EV71 and CA16 infections were previously reported to be distinct in terms of the interferon (IFN) response [15, 16]. In addition, in our previous studies, we observed an elevated expression of T-helper 2 (Th2) or Th1 cytokines in patients with EV71 or CA16 infection, respectively [17]. In parallel, as shown in experimental data on animals, an increase versus a decrease in immune-associated IFN-stimulated gene 15 (ISG15) expression was observed in animals with EV71 or CA16 infection, respectively [18]. To explore the potential rationale for the diversity in the immune response in humans infected by EV71 and CA16, our previous works analyzed the gene expression profiles in peripheral blood mononuclear cells (PBMCs) of rhesus macaques infected with these two viruses. The results showed that EV71 and CA16 infections could distinctively induce a wide variety of host genes, specifically, immune-associated genes such as interleukin (IL)-4 and IL-17 [19, 20].
Gene expression is usually precisely controlled by multiple biological events, such as pre-transcription (i.e., cis- and trans-acting regulators), post-transcription and translation [21, 22] events, as well as microRNAs (miRNAs) [23-26]. An miRNA is a non-coding, single-stranded, small RNA molecule of approximately 18–25 nucleotides. miRNAs are involved in the modulation of gene expression post-transcriptionally through either the degradation of target mRNA molecules or the repression of mRNA translation of the target gene [27-29]. Importantly, during viral infection, miRNAs are frequently observed as crucial effectors of events, such as viral replication and viral proliferation, by regulating cellular gene expression and altering regular cell function [29, 30]. And miRNAs are capable of not only playing a role in the innate immune response by modulating gene transcription and expression during viral infection [30-32] but also by regulating important genes associated with control of viral replication in the early phase of infection and genes associated with viral replication and packing in the middle phase.
To determine whether viral infection may induce a unbalance and abnormal immune response, we first focused on the miRNA expression profile induced in human fibroblasts (as a diploid cell originated from fetal lung tissue) infected with EV71 or CA16 and observed different characteristic miRNA profiles expressed in the cells infected by each respective virus. Further, the characterized miRNA molecules induced by each virus were verified and compared with the variation of some cytokines for Th1 and Th2 immune responses in infected rhesus macaques. These results seemingly suggest an interesting mechanism mediated by miRNA in the formation of specific immunity against EV71 or CA16 infection.
Materials & Methods
Viruses and cells
The EV71 virus (subgenotype C4) used in this study originated from an epidemic in Fuyang, China, in 2008 (GenBank accession no. EU812515.1) [33]. The CA16 virus G20 strain (subgenotype B) was isolated from a throat swab of an HFMD patient in Guangxi, China, in 2010 (GenBank accession no. JN590244.1) [34]. Both viruses proliferate in human diploid cells (human embryonic lung fibroblasts; Institute of Medical Biology (IMB), Chinese Academy of Medical Sciences (CAMS)). The human diploid cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (HyClone; Logan, UT, USA) with 10% fetal bovine serum (FBS) (Gibco; Grand Island, NY, USA).
Animal experiments
The animal experiments were designed based on the principles outlined in the Guide for the Care and Use of Laboratory Animals by the National Research Council of the National Academies [35] and the Guidance to Experimental Animal Welfare and Ethical Treatment by the Ministry of Science and Technology of the People’s Republic of China [36]. The experimental protocols were reviewed and approved by the Yunnan Provincial Experimental Animal Management Association (Approval no. SCXK (Dian) 2011-0005) and the Experimental Animal Ethics Committee of the Institute (Approval no. YISHENGLUNZI [2013] 4).
Two weeks before the experiments, no antibodies for either anti-EV71 or anti-CA16 were detected in any animal. Nine antibody-negative neonatal rhesus monkeys (40–50 days old) with weights ranging from 250 to 350 g were divided into three groups: three were in the EV71-infected group, three were in the CA16-infected group, and three were in the negative-control group (without infection). All the neonates were kept with their own mothers in single stainless-steel cages during the entire experimental period. The mothers were fed pellets (IMB, CAMS, China), peanuts and fresh fruit.
The experimental monkeys were infected with strain EV71 or CA16 (104.5 50% cell culture infective dose (CCID50) per animal) using a nasal spray (Zhang et al., 2014; Song et al., 2016), and the control animals were administered phosphate-buffered saline (PBS). Every day from the start of infection, the animals were monitored for clinical signs, and blood samples were collected under appropriate anesthesia to alleviate pain and minimize suffering, as described in the guidelines of IMB and CAMS. Following recovery from anesthesia after blood sample collection, all animals were returned to the colony with their mother.
Flow cytometry-based cytometric bead array analysis
Serum was collected from the monkeys to detect IL-2, IL-4, IL-5, IL-6, tumor necrosis factor-α (TNF-α) and interferon γ (IFN-γ). The assays used to detect these cytokines were conducted using a Th1/Th2 Cytokine Cytometric Bead Array Kit (BD Biosciences, San Diego, CA, USA) [37]. Briefly, a mixture of anti-cytokine beads was added to the serum samples, incubated with the phycoerythrin detection reagent (included in the kit) in the dark at room temperature for 3 h and then washed twice. The intensity of the resulting fluorescence signal was measured using a fluorescence-activated cell sorter flow cytometer (BD Biosciences), and the data were analyzed using cytometric bead array software (BD Biosciences).
Neutralizing antibody assay
The viral titer was analyzed by performing a microtitration assay using a standard protocol [38]. A mixture of diluted serum containing antibodies and the virus at a titer of 500–1000 CCID50 in 100 µL PBS was incubated at 37°C for 1 h. The cytopathic effect of the virus was examined by inoculating the mixture onto Vero cells grown in 96-well plates [39].
IFN-γ-specific enzyme-linked immunospot (ELISpot) assay
A standard ELISpot assay was performed as previously described [40]. Briefly, a 96-well polyvinylidene difluoride (PVDF)-backed plate was pre-coated with anti-IFN-γ monoclonal antibody by incubation overnight and blocked for 1 h at 37°C. Wells containing PBMCs of predetermined density and stimulating peptide (10 μg/mL) were incubated at 37°C for another 24 h. The cells were then removed, and the color was developed according to the manufacturer’s protocol. The colored spots were counted with an automated ELISpot reader (CTL, OH, USA). The spot-forming cells represented EV71 epitope-specific IFN-γ-producing T-cells.
Infection of human diploid cells in vitro
Human diploid cells were grown as a monolayer in DMEM with 10% FBS at 37 °C for 24 h and infected with the viral strains (multiplicity of infection = 1). The cells were harvested at 3 and 6.5 h post-infection (p.i.).
miRNA assay
Total miRNAs from the infected cells were collected using a mirVana miRNA Isolation Kit (Ambion; Austin, TX, USA). The RNA integrity number was also evaluated to determine RNA integrity using an Agilent Bioanalyzer 2100 (Agilent; Santa Clara, CA, USA). The extracted RNA was temporarily frozen in 95% ethanol at −20°C until further testing.
A human miRNA microarray (Agilent) was chosen to screen for miRNA expression in the infected cells. miRNA microarray experiments were conducted at the National Engineering Center for Biochip in Shanghai, China, according to the procedures in the Agilent technical manual. Briefly, miRNAs were labeled using a miRNA Completed Labeling and Hyb Kit (Agilent). Each slide was hybridized with 100 ng Cy3-labeled RNA using a miRNA Completed Labeling and Hyb Kit (Agilent) in a hybridization oven (Agilent). After hybridization, the slides were washed in staining dishes (Thermo; Waltham, MA, USA) with a Gene Expression Wash Buffer Kit (Agilent). Slides were scanned using a microarray scanner (Agilent), and the raw data were obtained using Feature Extraction software 10.7 (Agilent) and normalized using a quantile algorithm with Gene Spring 12.6 (Agilent). The systemic bioinformatic analyses of the microarray test were processed by Novel Bioinformatics Co., Ltd (Shanghai, China).
The differential expression of miRNAs was analyzed between in the CA16-infected and the EV71-infected group. Firstly, miRNA frequency was normalized to the number of transcripts per million clean tags (TPM). Then, the miRNAs whose expression changes greater than 3-fold or less than 0.15-fold were analyzed. A P-value < 0.05 was selected as the cut-off criterion. Target genes were predicted by the National Engineering Center for Biochip in Shanghai, China. The same target genes were used to draw the network.
Quantitative real-time polymerase chain reaction (qRT-PCR) confirmation
miRNAs were extracted from cells using a miRcute miRNA Isolation Kit (TIANGEN Bio; Beijing, China). The differential expression of four miRNAs, which were selected based on the miRNA assay, was confirmed by qRT-PCR using a miRcute Plus miRNA First-Strand cDNA Synthesis and qPCR Detection Kit (TIANGEN Bio) according to the manufacturer’s protocol. The primers for the miRNAs were provided by TIANGEN Bio.
Statistical analysis
The data were obtained in triplicate and are expressed as the mean values with standard deviation (SD). GraphPad Prism (San Diego, CA, USA) was used for the statistical analyses. The differences between two groups were evaluated using one-way analysis of variance. P< 0.05 was considered significant.
Results
miRNA expression profiling in human diploid cells infected with EV71 and CA16
Based on our data using microarrays showed extensive changes in immune-response-associated gene expression triggered by EV71 and CA16 infections [19, 20], and the fact that miRNAs usually plays a role in early or middle phase of viral infection , we screened all miRNAs to identify the major miRNAs associated with infection by these two viruses in vitro at 3 and 6.5 h p.i. The expression of 70 miRNAs changed at 3 h p.i in the EV71 infected human fibroblasts (as a diploid cell strain) compared with the uninfected controls (Table 1), comprising 43 upregulated miRNAs and 27 downregulated miRNAs (Table 1). This was more than in the CA16 infected cells at the same time point, which had only 28 differentially expressed miRNAs, 16 upregulated and 12 downregulated (Table 1). As shown in the miRNA expression profile of the human fibroblasts infected with EV71 and CA16, four miRNAs (miR-5585-3p, miR-619-5p, miR-6786-5p and miR-8071) exhibited wide variations in expression during both EV71 and CA16 infection (Figure 1). Among these miRNAs, compared with the negative-control (uninfected) group, the levels of miR-5585-3p and miR-619-5p decreased after viral infection, and the level of miR-6786-5p increased after viral infection (Figure 1). In contrast, the level of miR-8071 tended to be up-regulated in the CA16-infected group, whereas the same miRNA was up-regulated (at 3 h p.i.) and down-regulated (at 6.5 h p.i.) in the EV71-infected group (Figure 1). Thus, these target genes may be involved in viral identification (such as receptors and stress responses) and in antiviral signaling pathways (Table 2).
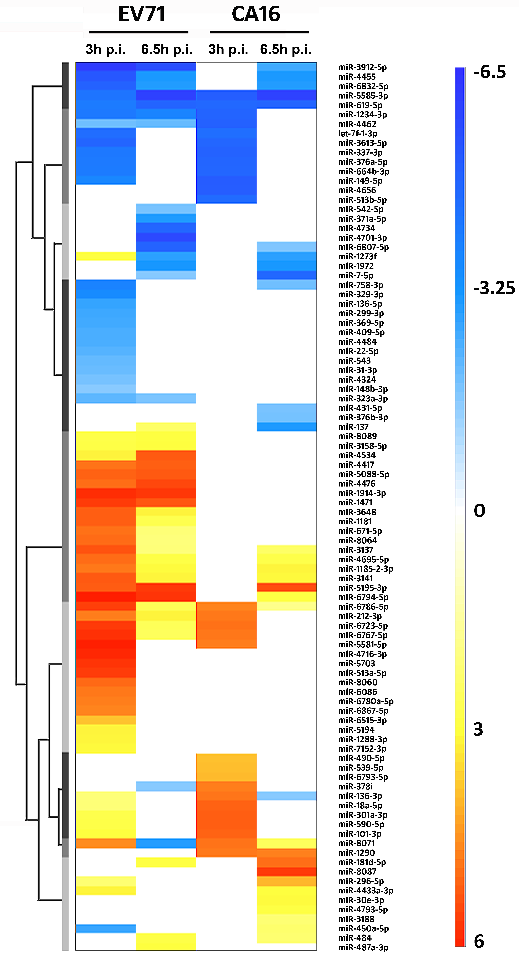
Figure 1. miRNA expression profiles of human diploid cells infected with EV71 and CA16 in vitro.
Total miRNAs from infected cells (multiplicity of infection = 1) were extracted for miRNA assay at 3 and 6.5 h p.i. The heat map of the miRNA expression profile was normalized to that of the negative-control group (uninfected group). Down-regulated miRNAs are shown in blue and up-regulated miRNAs are shown in red. The values are shown on a log2 scale.
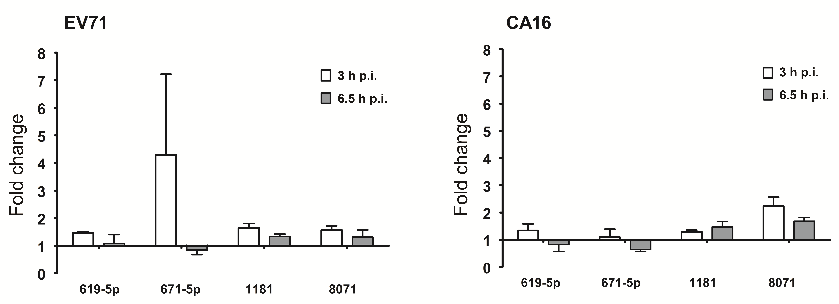
Figure 2. Confirmation of miRNA expression by qRT-PCR. qRT-PCR analysis of four selected miRNAs expressed after EV71 and CA16 infection at 3 and 6.5 h p.i.
The results were normalized to the level of the small nuclear RNA U6, which is widely used endogenous reference RNA in miRNA quantification studies. The y-axis indicates the relative quantity of the specific miRNA in the infected samples compared with the uninfected samples. Error bars indicate the SD of the relative quantities.
Table 1. Summary of differentially expressed miRNAs induced by EV71 and CA16 infections.
|
Virus |
3 h p.i. |
|
6.5 h p.i. |
||||
|
|
Total |
Up-regulated |
Down-regulated |
|
Total |
Up-regulated |
Down-regulated |
|
EV71 |
70 |
43 |
27 |
|
44 |
26 |
18 |
|
CA16 |
28 |
16 |
12 |
|
32 |
18 |
14 |
The number of differentially expressed miRNAs is shown. The levels of miRNAs induced by EV71 and CA16 were normalized to those of the negative-control group (uninfected group).
Table 2. Common miRNAs induced by EV71 and CA16 infection in human diploid cells.
|
miRNA |
EV71 |
CA16 |
Predicted target gene |
Description |
||
|
3hp.i. |
6.5hp.i. |
3hp.i. |
6.5hp.i. |
|||
|
hsa-miR-5585-3p |
0.05 |
0.02 |
0.03 |
0.02 |
XPR1 |
Virus receptor activity |
|
|
|
|
|
|
DNAJB7 |
Heat shock protein (Hsp40) |
|
hsa-miR-619-5p |
0.06 |
0.03 |
0.04 |
0.03 |
TIAF1 |
Activated Th cells |
|
hsa-miR-6786-5p |
46.86 |
6.32 |
25.07 |
3.51 |
NA |
NA |
|
hsa-miR-8071 |
23.10 |
0.12 |
29.03 |
5.87 |
PPP2R1A |
Regulation of Wnt signaling pathway |
NA, not applicable; there is no predicted target.
Numbers indicate fold change, normalized to the negative-control group (uninfected group).
In addition to these four miRNAs that changed during both EV71 and CA16 infections, there were many other miRNAs induced only by EV71 or CA16 infection (Figure 1). In general, there were more changes in miRNAs induced by EV71 infection: miR-4417, miR-5088-5p, miR-4476, miR-1914-3p and miR-1471 were all up-regulated at both 3 and 6 h p.i., whereas no significant alterations were observed with CA16 infection for these miRNAs (Figure 1). Similarly, EV71 infection induced remarkable down-regulation of miR-323a-3p at two different time points, but CA16 infection did not induce any alteration of this miRNA (Figure 1). To confirm the results obtained from microarray analysis, qRT-PCR was performed using primers designed based on the sequences of the miRNAs. A total of four miRNAs were selected for primer design. The miRNA expression quantified by qRT-PCR showed the same patterns as those observed in the miRNA assay (Figure 2). The target genes of these miRNAs, which exhibited remarkable variations in expression, were found to be involved in immune response, signal transduction and transcriptional regulation (Table 3 and Figure 3). All these findings suggest that EV71 and CA16 infections could induce specific immune responses via individual signaling pathways and modulatory approaches.
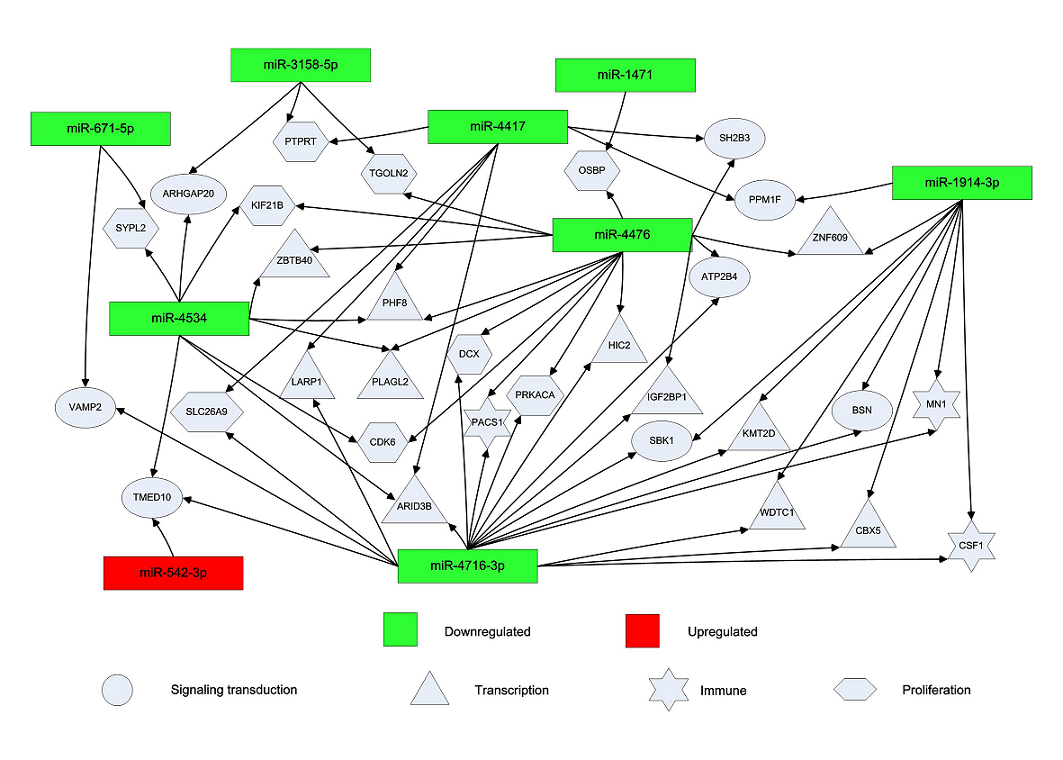
Figure 3. Network of differentially expressed miRNAs and their target genes during EV71 and CA16 infection.
The levels of miRNAs induced by CA16 were normalized to those of the EV71-infected group. Fold changes greater than 3-fold or less than0.15-fold are shown. Down-regulated miRNAs are shown in green and up-regulated miRNAs in red. Genes involved in signal transduction are indicated by ellipses, genes involved in transcription by triangles, immune response-related genes by stars and proliferation-related genes by polygons.
Table 3. Differentially expressed miRNAs induced by EV71 and CA16.
|
miRNA |
EV71 |
CA16 |
Predicted target gene |
Description |
||
|
3 h p.i. |
6.5 h p.i. |
3 h p.i. |
6.5 h p.i. |
|
|
|
|
hsa-miR-323a-3p |
0.21 |
0.29 |
NA |
NA |
IGSF3 |
Immune |
|
|
|
|
|
|
UBE2K |
Ubiquitin |
|
hsa-miR-671-5p |
29.57 |
4.34 |
NA |
NA |
ZNF318 |
Transcription |
|
|
|
|
|
|
RAB6B |
Signaling transduction |
|
|
|
|
|
|
HIVEP3 |
Transcription (promotes TNF-α-induced apoptosis) |
|
|
|
|
|
|
CDIP |
Transcription (regulates TNF-α-induced apoptosis by p53) |
|
hsa-miR-1181 |
35.34 |
6.72 |
NA |
NA |
SOX2 |
Transcription (negative regulation of Wnt signaling pathway) |
|
|
|
|
|
|
STAT3 |
Signaling transduction (is activated by IFNs, IL-5, IL-6, etc.) |
|
hsa-miR-1290 |
NA |
NA |
27.42 |
25.68 |
MSR1 |
Immune |
|
|
|
|
|
|
ZFHX3, |
Transcription |
|
|
|
|
|
|
RGS7BP |
Signaling transduction |
|
hsa-miR-1471 |
48.49 |
37.76 |
NA |
NA |
OSBP |
Transporter (positive regulation of growth of symbiont in host, i.e.,hepatitis C virus, poliovirus) |
|
hsa-miR-1914-3p |
55.37 |
50.22 |
NA |
NA |
TCF7 |
Immune (regulates lymphocyte differentiation) |
|
|
|
|
|
|
E2F1 |
Transcription |
|
|
|
|
|
|
HDAC1 |
Transcription |
|
hsa-miR-3158-5p |
7.39 |
8.04 |
NA |
NA |
CNOT7 |
Immune (B-cell translocation, anti-proliferation) |
|
|
|
|
|
|
GABBR2 |
Signaling transduction |
|
|
|
|
|
|
CSNK1G1 |
Metabolism |
|
hsa-miR-3648 |
35.22 |
8.97 |
NA |
NA |
APC2 |
Signaling transduction (negative regulation of Wnt signaling pathway) |
|
hsa-miR-4417 |
29.03 |
34.88 |
NA |
NA |
NFATC3 |
Transcription (regulates the gene expression in T- cells and immature thymocytes) |
|
|
|
|
|
|
UBE2D2 |
Ubiquitin |
|
|
|
|
|
|
TFAM |
Transcription |
|
hsa-miR-4476 |
30.69 |
43.30 |
NA |
NA |
PBX1, |
Transcription |
|
|
|
|
|
|
XPO5 |
Transporter (transports small RNAs and double-stranded RNA-binding proteins from the nucleus to the cytoplasm) |
|
hsa-miR-4534 |
8.92 |
36.63 |
NA |
NA |
LTBP2 |
Signaling transduction |
|
|
|
|
|
|
DOCK3 |
Signaling transduction (negative regulation of Wnt signaling pathway) |
|
hsa-miR-8064 |
31.25 |
4.19 |
NA |
NA |
OR10G4 |
Signaling transduction |
|
|
|
|
|
|
SLC6A4 |
Transporter (transports neurotransmitter) |
|
hsa-miR-8089 |
7.20 |
7.87 |
NA |
NA |
CXCL14 |
Immune (chemokine) |
|
|
|
|
|
|
GABRG3 |
Signaling transduction |
NA, not applicable.
Numbers indicate fold change, normalized to the negative-control group (uninfected group).
Immune response-associated miRNA expression profiling in human diploid cells infected with EV71 and CA16
Nevertheless, based upon our above observation, CA16 infection still failed to induce a miRNA expression profile similar to that induced by EV71 infection in its variety or dynamic range (Figure 1, Table 1 and Figure 2). Therefore, we further analyzed the immune response-associated miRNA expression profile induced by EV71 and CA16 infections to explore potential clues regarding the diversity of their pathogenicity. Unlike EV71 infection, CA16 infection directly led to the general down-regulation of miRNAs whose target genes are associated with negative regulation of the immune response (Figure 4). These data suggest that miRNAs may be used to suppress the immune response during CA16 infection.
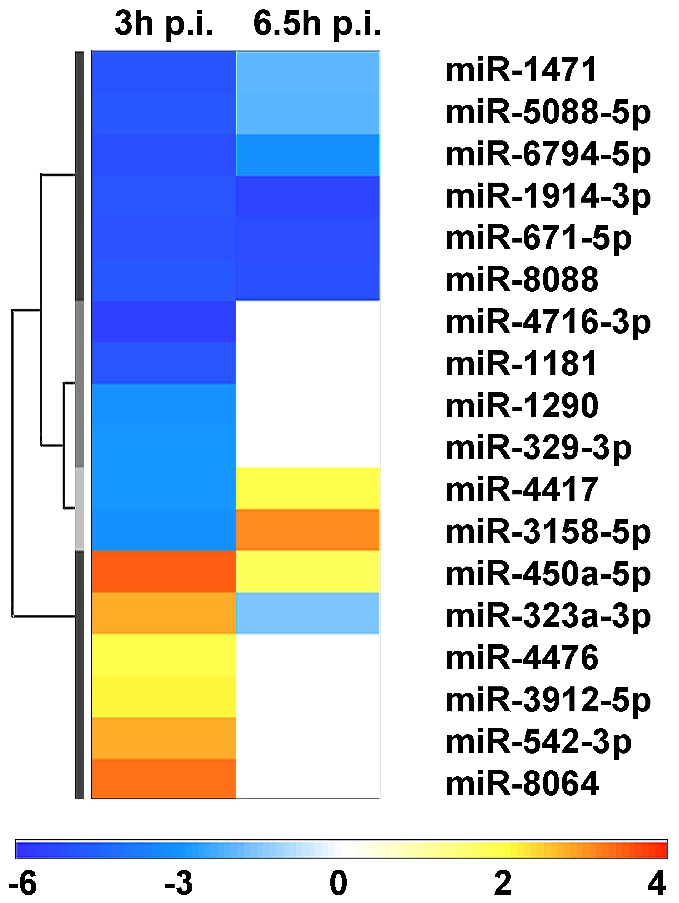
Figure 4. Differences in miRNAs associated with the immune response between EV71 and CA16 infection.
The levels of miRNAs induced by CA16 were normalized to those of the EV71-infected group. Down-regulated miRNAs are shown in blue and up-regulated miRNAs in red. The values are shown on a log2 scale.
Partial cytokine expression in rhesus macaques infected with EV71 and CA16
Based upon our observation of miRNA expression profiles induced in human diploid cells by EV71 and CA16 described above, we tried to link the variation of miRNA expression profile characterized by EV71 versus CA16 in cultured cells to the immune responses observed in primates infected by the two respective viruses. As in a previous study [41], vesicles in the mouth and on the limbs were observed in the infected rhesus macaques (Figure 5A). Varied expression of released cytokines was detected in the peripheral blood of rhesus macaques infected with EV71 or CA16, and the results showed wide diversity in the levels of partial cytokines (e.g., IL-5, TNF-α and IFN-γ) (Figure 5B). In EV71-infected rhesus macaques, the levels of IL-5, IL-6, TNF-α and IFN-γ were markedly increased and typically sustained for a period of time (Figure 5B); in contrast, in CA16-infected rhesus macaques, the levels of IL-6 and TNF-α were transiently elevated on day 7 p.i.; however, no significant changes were noted in the other cytokines (Figure 5B).
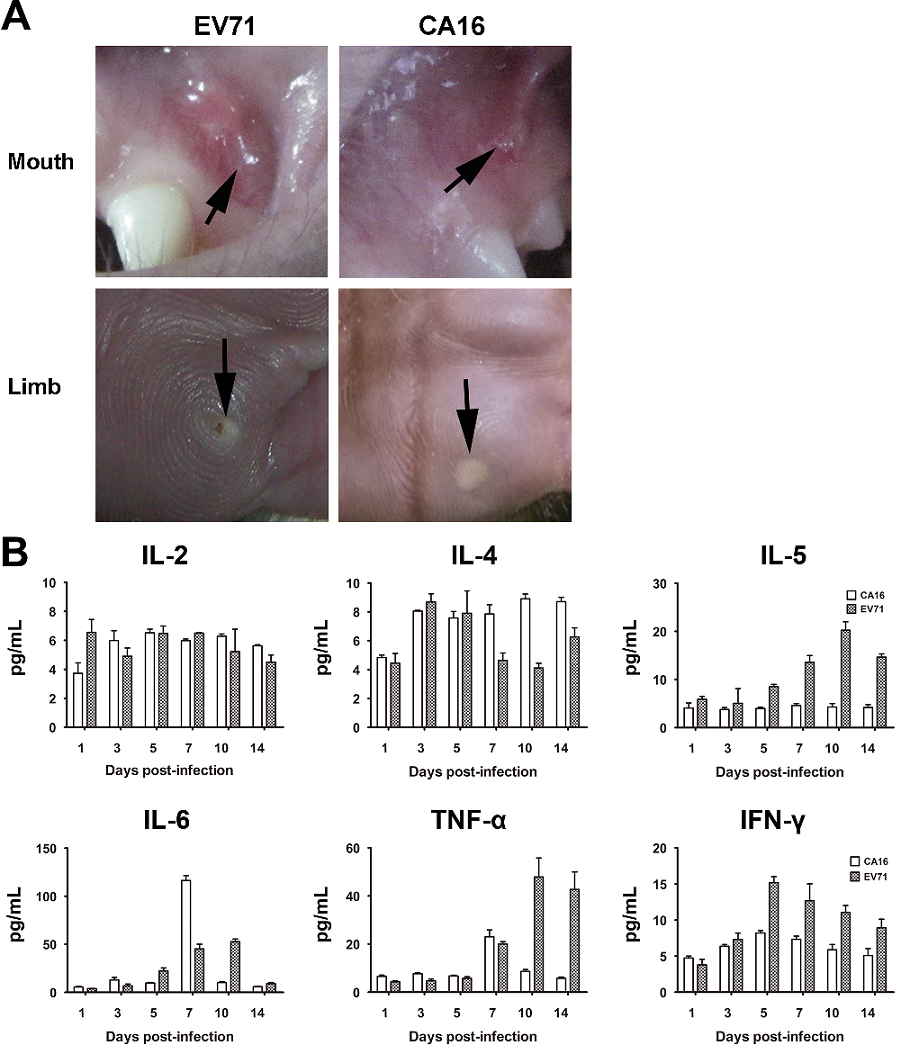
Figure 5. Cytokines released in rhesus macaques infected with EV71 or CA16.
Serum from the peripheral blood of rhesus macaques infected with EV71 or CA16 was obtained on days 1, 3, 5, 7, 10 and 14 p.i. and a cytokine as saw as conducted.
Immune response in rhesus macaques infected with EV71 or CA16
To examine the immune responses induced by EV71 and CA16 infections, we compared the humoral response (i.e., neutralizing antibodies) and the cellular response (i.e., IFN-γ) in rhesus macaques infected with EV71 or CA16. Unlike the robust anti-EV71-specific humoral and cellular responses induced by EV71 infection [42] (Figure 6), the anti-CA16 neutralizing antibody titer induced by CA16 infection was lower than 1:4 (identified as negative according to the standard). There was no difference between the IFN-γ-specific cellular response induced in PBMCs infected with CA16 and that in the negative-control macaques (normal macaques) (Figure 6).
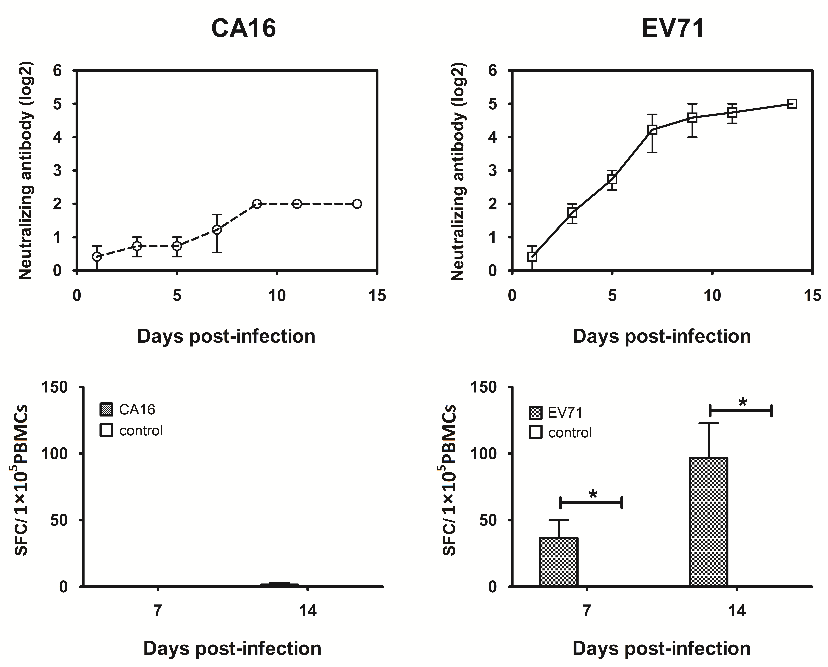
Figure 6. Neutralizing antibody and IFN-γ induced by EV71 and CA16 infection in rhesus macaques.
Serum from the peripheral blood of rhesus macaques infected with EV71 and CA16 was obtained to measure the levels of neutralizing antibody on days 0, 3, 5, 7, 9, 11 and 14 p.i. In addition, PBMCs from the peripheral blood sample of these macaques were obtained to measure the levels of IFN-γ on days 0, 7 and 14 after infection. SFC: spot-forming cell. *: p<0.05.
Discussion
Most previous studies have typically focused on analysis of the pathological and immunological responses resulting from EV71 or CA16 infection [10, 41, 43]. Leng and colleagues reported that EV71 infection activates the immune response via natural killer T-cells but that CA16 activates the immune response via type I IFN [15, 16]. However, the data comparing the differences between these two viral infections in detail are scarce. To explore the characteristics of the immune response during EV71 and CA16 infections, we systematically analyzed the expression of miRNAs induced by EV71 and CA16 in cultured human diploid cells in the current study, and the results of the miRNA expression profile obtained from the infected cells were verified further based upon the variation of some functional molecules associated with the immune response detected in the macaques infected by both viruses. The consistency in this verification showed a trend similar to that observed in a comparison of gene expression profiles and immunologic factors in blood induced by both viruses in infected macaques [19, 20].
In the current work, the first important observation was that miRNAs associated with the recognition of viral pathogen-associated molecular patterns (PAMPs) and antiviral signal transduction pathways increased during both EV71 and CA16 infections, as with hepatitis C (HCV) and tobacco mosaic virus (TMV) [44-46]. These findings suggest that the human diploid cells, as fibroblasts originated from lung epithelium tissue, are capable of responding to viral PAMPs through pattern recognition receptors (PRRs) and could logically release signal molecules to activate the host innate immune response. In this case, a marked difference in the expression levels of miRNAs associated with signal transduction and transcriptional regulation in cells infected with EV71 or CA16 may suggest the different interaction of fibroblasts with EV71 and CA16, which may lead to various stimulations to the process of adaptive immune response formation during viral infection. Compared with EV71 infection, fewer miRNAs were induced by CA16 for a normal immune response against viral antigens. Particularly in cells infected with CA16, the levels of miR-5088-5p were reduced. One of the target genes for miR-5088-5p is AKT2, which is involved in the negative modulation of the B-/T-cell response, resulting in failure to activate the release of immune-associated cytokines and immune response repression; the same effect was observed for miR-6794-5p. Transient elevation of the levels of several cytokines (IL-6 and TNF-α) has been observed in CA16-infected rhesus macaques within the period of acute infection [17, 41, 43], and neutralizing antibody and IFN-γ were not induced by CA16. It suggests that CA16 infection is unable to cause a rapid and intense immune response compared with EV71 infection, which seems to suggest an inhibiting effect during formation of the immune response to CA16 infection. These results are consistent with a report by Luo et al. [47], who suggested that CA16 infection stimulates imbalances of T-cells. Notably, it is almost impossible for hosts to protect against recurrent CA16 infection due to the lack of previously characterized immune response indicators [7, 8]. According to these results, we speculate that miRNAs may play an important role involved in the regulation of immune responses against CA16 infection. miRNA molecules, which involved in regulating the T-/B-cell activation response to CA16 infection, were reduced in cells. In contrast, EV71 infection could simply activate the regular B-/T-cell response due to the failure to downregulate the expression of negatively controlled miRNAs, thereby eliciting effective humoral immunity and specific cellular responses that protect hosts from recurrent infection by EV71 virus in clinical practice.
Nevertheless, the miRNAs we discovered are not the same miRNAs that were previously reported by Cui et al. [48]. We are unable to determine whether these two studies can be compared because the miRNA expression profiling from our study was induced in cultured cells infected by EV71 and CA16 in vitro, but the former report used the serum of patients infected by these two viruses. In addition, in our study, detection of miRNA started from the early phase of viral infection (3 and 6.5 h), and detection of cytokines was carried out over the whole acute viral infection in infected animals (14 days), which was predicted to provide data regarding the variation of some cytokines associated with viremia. In contrast, Cui et al. [48] only analyzed patients starting from when symptoms appeared, mainly during the onset phase (usually 3–7 days after viral infection). Thus, differences between the two studies are understandable.
Interestingly, in our study, the expression levels of these immune effectors induced by EV71 and CA16 infection in macaques were similar to levels of the associated cytokines in patients infected with the two viruses [17, 19, 20]. It prompts that macaque might be a candidate animal model to research about EV71 and CA16 infection.
Absolutely, there are some limits in this study. Firstly, the differentially expressed miRNAs should be confirmed by further experiments. Secondly, the more details of mechanism for regulation immune response by these important miRNAs should be revealed in further studies.
Conclusion
Based on these results and our previous research, which described the acute infectious process of EV71 and CA16 in rhesus monkeys and analyzed Th1 and Th2 immune responses induced during these infections [17, 41, 43, 49], the differences in miRNA profiles induced by these two viruses in cells obtained here may provide clues for further investigations into the relationship of these viruses with the immune response and the role of miRNAs in immune regulation.
Acknowledgement
Author’s contributions
Conceived and designed the experiments: QHL, YG, YZ. Performed the experiments: YG, YZ, JBW, JJW, XLX, YouG, YL. Analyzed the data: QHL, YG, YZ. Contributed reagents/materials/analysis tools: YCC, MF, STF, ZFY, XQD. Wrote the paper: QHL, YZ. The manuscript was drafted by QHL.
Funding
This work was supported by CAMS Initiative for Innovative Medicine (CAMS-I2M-1-019), the State Project for Essential Drug Research and Development (2017ZX09307013-2 and 2016ZX09101120-2), the National Natural Science Foundation (31700931), Medical reserve talents of Yunnan Provincial Health and Family Planning (H-201620) and Major science and technology special projects of Yunnan Province (2017ZF020, 2016ZF002, 2014HB066, 2017FB018).
References
Yang F, Ren L, Xiong Z, Li J, Xiao Y, Zhao R, et al. Enterovirus 71 outbreak in the People's Republic of China in 2008. J Clin Microbiol 2009;47(7):2351-2. PMid:19439545
View Article PubMed/NCBILi L, He Y, Yang H, Zhu J, Xu X, Dong J, et al. Genetic characteristics of human enterovirus 71 and coxsackievirus A16 circulating from 1999 to 2004 in Shenzhen, People's Republic of China. J Clin Microbiol 2005;43(8):3835-9. Mid:16081920
View ArticleSuzuki Y, Taya K, Nakashima K, Ohyama T, Kobayashi JM, Ohkusa Y, et al. Risk factors for severe hand foot and mouth disease. Pediatr Int 2010;52(2):203-7. PMid:19663940
View Article PubMed/NCBIChang LY, Lin TY, Hsu KH, Huang YC, Lin KL, Hsueh C, et al. Clinical features and risk factors of pulmonary oedema after enterovirus-71-related hand, foot, and mouth disease. Lancet 1999;354(9191):1682-6. 04434-7
View ArticleHo M, Chen ER, Hsu KH, Twu SJ, Chen KT, Tsai SF, et al. An epidemic of enterovirus 71 infection in Taiwan. Taiwan Enterovirus Epidemic Working Group. N Engl J Med 1999;341(13):929-35. PMid:10498487
View Article PubMed/NCBIKomatsu H, Shimizu Y, Takeuchi Y, Ishiko H, Takada H. Outbreak of severe neurologic involvement associated with Enterovirus 71 infection. Pediatr Neurol 1999;20(1):17-23. 00087-3
View ArticleChen GP, Wu JB, Wang JJ, Pan HF, Zhang J, Shi YL, et al. Epidemiological characteristics and influential factors of hand, foot and mouth disease (HFMD) reinfection in children in Anhui province. Epidemiol Infect 2016;144(1):153-60. PMid:26027435
View Article PubMed/NCBITagaya I, Tachibana K. Epidemic of hand, foot and mouth disease in Japan, 1972-1973: difference in epidemiologic and virologic features from the previous one. Jpn J Med Sci Biol 1975;28(4):231-4. PMid:175202
View Article PubMed/NCBIWu Z, Gao Y, Sun L, Tien P, Jin Q. Quick identification of effective small interfering RNAs that inhibit the replication of coxsackievirus A16. Antiviral Res 2008;80(3):295-301. PMid:18662724
View Article PubMed/NCBIMao Q, Wang Y, Gao R, Shao J, Yao X, Lang S, et al. A neonatal mouse model of coxsackievirus A16 for vaccine evaluation. J Virol 2012;86(22):11967-76. PMid:22951825
View Article PubMed/NCBIZhu G, Zheng Y, Zhang L, Shi Y, Li W, Liu Z, et al. Coxsackievirus A16 infection triggers apoptosis in RD cells by inducing ER stress. Biochem Biophys Res Commun 2013;441(4):856-61. PMid:24211204
View Article PubMed/NCBILi JP, Liao Y, Zhang Y, Wang JJ, Wang LC, Feng K, et al. Experimental infection of tree shrews (Tupaia belangeri) with Coxsackie virus A16. Dongwuxue Yanjiu 2014;35(6):485-91. PMid:25465084
PubMed/NCBISingh S, Chow VT, Chan KP, Ling AE, Poh CL. RT-PCR, nucleotide, amino acid and phylogenetic analyses of enterovirus type 71 strains from Asia. J Virol Methods 2000;88(2):193-204. 00185-3
View ArticleSolomon T, Lewthwaite P, Perera D, Cardosa MJ, McMinn P, Ooi MH. Virology, epidemiology, pathogenesis, and control of enterovirus 71. Lancet Infect Dis 2010;10(11):778-90. 70194-8
View ArticleYang J, Yang C, Guo N, Zhu K, Luo K, Zhang N, et al. Type I Interferons Triggered through the Toll-Like Receptor 3-TRIF Pathway Control Coxsackievirus A16 Infection in Young Mice. J Virol 2015;89(21):10860-7. PMid:26292317
View Article PubMed/NCBIZhu K, Yang J, Luo K, Yang C, Zhang N, Xu R, et al. TLR3 signaling in macrophages is indispensable for the protective immunity of invariant natural killer T cells against enterovirus 71 infection. PLoS Pathog 2015;11(1):e1004613. PMid:25615690
View Article PubMed/NCBIWang J, Pu J, Liu L, Che Y, Liao Y, Wang L, et al. Clinical and Associated Immunological Manifestations of HFMD Caused by Different Viral Infections in Children. Global Pediatric Health 2016;3:1-9. PMid:27336013
View Article PubMed/NCBIChan SY, Sam IC, Lai JK, Chan YF. Cellular proteome alterations in response to enterovirus 71 and coxsackievirus A16 infections in neuronal and intestinal cell lines. J Proteomics 2015;125:121-30. PMid:26003530
View Article PubMed/NCBIZhang Y, Yang E, Pu J, Liu L, Che Y, Wang J, et al. The gene expression profile of peripheral blood mononuclear cells from EV71-infected rhesus infants and the significance in viral pathogenesis. PLoS One 2014;9(1):e83766. PMid:24392094
View Article PubMed/NCBISong J, Hu Y, Hu Y, Wang J, Zhang X, Wang L, et al. Global gene expression analysis of peripheral blood mononuclear cells in rhesus monkey infants with CA16 infection-induced HFMD. Virus Res 2016;214:1-10. PMid:26775814
View Article PubMed/NCBIXu S, Wu D. Effect and regulation of fatty acid in gene expression. Feed Industry 2007;28(5):16-19.
Trivedi MS, Deth RC. Role of a redox-based methylation switch in mRNA life cycle (pre- and post-transcriptional maturation) and protein turnover: implications in neurological disorders. Front Neurosci 2012;6:92. PMid:22740813
View Article PubMed/NCBIHamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 1999;286(5441):950-2.
View ArticleWaterhouse PM, Wang MB, Finnegan EJ. Role of short RNAs in gene silencing. Trends Plant Sci 2001;6(7):297-301. 01989-6
View ArticleCarrington JC, Ambros V. Role of microRNAs in plant and animal development. Science 2003;301(5631):336-8. PMid:12869753
View Article PubMed/NCBICheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res 2005;33(4):1290-7. PMid:15741182
View Article PubMed/NCBIBartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 2004;5(5):396-400. PMid:15143321
View Article PubMed/NCBIBartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 2004;116(2):281-97. 00045-5
View ArticleHe L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 2004;5(7):522-31. PMid:15211354
View Article PubMed/NCBIBaltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol 2008;9(8):839-45. PMid:18645592
View Article PubMed/NCBIGantier MP, Sadler AJ, Williams BR. Fine-tuning of the innate immune response by microRNAs. Immunol Cell Biol 2007;85(6):458-62. PMid:17621315
View Article PubMed/NCBIRoy S, Sen CK. MiRNA in innate immune responses: novel players in wound inflammation. Physiol Genomics 2011;43(10):557-65. PMid:21139022
View Article PubMed/NCBIMa S-h, Liu J-s, Wang J-j, Shi H-j, Yang H-j, Chen J-y, et al. Genetic Analysis of the VP1 Region of Human Enterovirus 71 Strains Isolated in Fuyang, China, During 2008. Virologica Sinica 2009;24(3):162-70.
View ArticleYang E, Cheng C, Zhang Y, Wang J, Che Y, Pu J, et al. Comparative study of the immunogenicity in mice and monkeys of an inactivated CA16 vaccine made from a human diploid cell line. Hum Vaccin Immunother 2014;10(5):1266-73. PMid:24583556 PMCid:PMC4896597
View Article PubMed/NCBILab_Animal. Guide for the Care and Use of Laboratory Animals. 8th Ed ed. Washington, DC: National Academies Press, 2011.
Republic_of_China TMoSaTotPs. The Guidance to experimental animal welfare and ethical treatment. In, 2006.
Chen R, Lowe L, Wilson JD, Crowther E, Tzeggai K, Bishop JE, et al. Simultaneous Quantification of Six Human Cytokines in a Single Sample Using Microparticle-based Flow Cytometric Technology. Clin Chem 1999;45(9):1693-94. PMid:10471693
PubMed/NCBIArita M, Nagata N, Sata T, Miyamura T, Shimizu H. Quantitative analysis of poliomyelitis-like paralysis in mice induced by a poliovirus replicon. J Gen Virol 2006;87(Pt 11):3317-27. PMid:17030866
View Article PubMed/NCBIWHO. Procedure for Using the Lyophilized LBM Pools for Typing Enterovirus, Geneva. 1988.
Streeck H, Frahm N, Walker BD. The role of IFN-gamma Elispot assay in HIV vaccine research. Nat Protoc 2009;4(4):461-9. PMid:19282851
View Article PubMed/NCBILiu L, Zhao H, Zhang Y, Wang J, Che Y, Dong C, et al. Neonatal rhesus monkey is a potential animal model for studying pathogenesis of EV71 infection. Virology 2011;412(1):91-100. PMid:21262515
View Article PubMed/NCBIDong C, Liu L, Zhao H, Wang J, Liao Y, Zhang X, et al. Immunoprotection elicited by an enterovirus type 71 experimental inactivated vaccine in mice and rhesus monkeys. Vaccine 2011;29(37):6269-75. PMid:21722686
View Article PubMed/NCBIZhang Y, Cui W, Liu L, Wang J, Zhao H, Liao Y, et al. Pathogenesis study of enterovirus 71 infection in rhesus monkeys. Lab Invest 2011;91(9):1337-50. PMid:21555996
View Article PubMed/NCBILi Y, Zhang Q, Zhang J, Wu L, Qi Y, Zhou JM. Identification of microRNAs involved in pathogen-associated molecular pattern-triggered plant innate immunity. Plant Physiol 2010;152(4):2222-31. PMid:20164210
View Article PubMed/NCBIThomas E, Gonzalez VD, Li Q, Modi AA, Chen W, Noureddin M, et al. HCV infection induces a unique hepatic innate immune response associated with robust production of type III interferons. Gastroenterology 2012;142(4):978-88. PMid:22248663
View Article PubMed/NCBIBoccara M, Sarazin A, Thiebeauld O, Jay F, Voinnet O, Navarro L, et al. The Arabidopsis miR472-RDR6 silencing pathway modulates PAMP- and effector-triggered immunity through the post-transcriptional control of disease resistance genes. PLoS Pathog 2014;10(1):e1003883. PMid:24453975
View Article PubMed/NCBILuo Q, Peng W, Chen LI. Coxsackievirus A16 infection stimulates imbalances of T cells in children. Exp Ther Med 2015;9(6):2213-18. PMid:26136962
View Article PubMed/NCBICui L, Qi Y, Li H, Ge Y, Zhao K, Qi X, et al. Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease. PLoS One 2011;6(11):e27071. PMid:22087245
View Article PubMed/NCBILiang Y, Zhou X, Yang E, Pu J, Che Y, Wang J, et al. Analysis of the Th1/Th2 reaction in the immune response induced by EV71 inactivated vaccine in neonatal rhesus monkeys. J Clin Immunol 2012;32(5):1048-58. PMid:22585051
View Article PubMed/NCBI
