Toshihiko Hanai
Phone: 81-45-547-4871; E-mail: thanai@kf7.so-net.ne.jp
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 2 ISSUE: 3
Page No: 190-198
Toshihiko Hanai
Phone: 81-45-547-4871; E-mail: thanai@kf7.so-net.ne.jp
Toshihiko Hanai1,2*, Masumi Katane2, Hiroshi Homma2
Izuru Kawamura(izuruk@ynu.ac.jp)
Silvia Sacchi(silvia.sacchi@uninsubria.it)
Toshihiko Hanai, Quantitative in silico analysis of selective enzyme reaction of mammalian D-amino acid oxidase and acidic D-amino acid oxidase mutants(2018)SDRP Journal of Computational Chemistry & Molecular Modelling 2(3)p:190-198
The mammalian D-amino acid oxidase (DAO) and D-aspartate oxidase (DDO) are presumed to regulate the levels of several endogenous and exogenous D-amino acids in various organs; however, their physiological roles in vivo remain to be fully clarified. The oxidation reactivity of DAO and DDO was quantitatively analyzed in silico.
The stereo structure of mammalian DAO was constructed in silico from pig kidney DAO (PDB ID: 1VE9) and optimized using the MM2 calculation, then, atomic partial charge (apc) of key atoms was calculated using a MOPAC-PM5 program, and the specific activity (U mg-1 protein) for several substrate amino acids was correlated with the change of apc values (Δapc) of α-carbon (αC) of the amino acids. The feasibility of the constructed mammalian DAO was analyzed by the comparison with four downloaded mammalian DAOs. Furthermore, the stereo structures of mouse DDO was constructed in silico from pig DAO and optimized using MM2 calculation, and apc of key atoms was calculated using MOPAC-PM5 program, and then correlated with the oxidation reactivity. Likewise, the DDO mutants carrying a substitution of alanine, glycine, or tyrosine for the active site residue serine-308 (S308A, S308G, and S308Y mutant, respectively) and a double-point substitution of glycine and tyrosine for serine-308 and glycine-309, respectively (S308G-G309Y mutant), were also analyzed.
Correlation coefficient between the specific activity (U mg-1 protein) of the constructed mammalian DAO, downloaded mammalian DAO (PDB ID: 2du8, 3g3e, and 3znn) and apc of αC of substrate was over 0.8 (n = 14). Furthermore, the reactivity (Vmax/Km) of mouse DDO and DDO mutants was correlated with Δapc of αC and αN of D-aspartic acid with r = 0.849 (n = 5) and r = 0.876 (n = 5), and with that of those of NMDA with r = 0.902 (n = 5) and r = 0.741 (n = 5), respectively. Conformation analysis of DAO complex with several active site-directed inhibitors did not quantitatively demonstrate the enzyme reactivity. The reactivity can be quantitatively described by electron localization of key atoms. The apc of key atoms can support the oxidation reaction of DAO and DDO. The method described herein may contribute to basic enzymology and application to the development of useful enzyme inhibitors.
KEYWORDS: Acidic D-amino acid oxidase mutant, Atomic partial charge, Mammalian D-amino acid oxidase, Molecular mechanics, Quantitative in silico analysis, Selective enzyme reaction,
Quantitative analysis of enzyme reactivity is a basic requirement for the development of precise diagnostic methods and drug discovery processes, and reduces the number of animal tests. Designed engineering proteins are preferred for developing effective synthesis methods for industrial uses. The specific affinity for an enzyme reaction can be analyzed in silico, but it is difficult to develop a general rule for the selectivity of the enzyme reaction because of a lack of sufficient data. Developing computational chemical analyses have enabled the structure-based in silico screening of many compounds listed in chemical structure databases. However, the enzyme reactivity has not been quantitatively analyzed.
The oxidation reactivity of D-amino acid oxidase (DAO) was quantitatively analyzed in silico [1-3], because D-amino acids exist in manmal’s tissues, especially in brain [4-6]. DAO is the catabolic enzyme that stereo specifically acts on several neutral and basic D-amino acids (but not on acidic D-amino acids), including free D-serine [7], which plays an important role in regulating brain function by acting as an endogenous ligand for the N-methyl-D-aspartate (NMDA) receptor (a subtype of the L-glutamate receptor family) and α d2 glutamate receptor [8-10]. Besides D-serine, free D-aspartic acid has also been studied extensively. Unlike the tissue (brain)-specific expression of D-serine, substantial amounts of D-aspartic acid are present in a wide variety of tissues and cells in mammals, particularly those that make up the central nervous, neuroendocrine, and endocrine systems. Several lines of evidence suggest that D-aspartic acid plays an important role in regulating developmental processes, hormone secretion, and steroidogenesis [11, 12]. Acidic D-amino acids, such as D-aspartic acid, D-glutamic acid, and NMDA are degraded by another D-amino acid-specific enzyme, D-aspartate oxidase (DDO). DAO and DDO are flavin adenine dinucleotide (FAD)-containing flavoproteins that catalyze the oxidative deamination of D-amino acids to generate 2-oxo acids along with hydrogen peroxide and ammonia. The mammalian DAO and DDO are presumed to regulate the levels of several endogenous and exogenous D-amino acids, including D-serine and D-aspartic acid, in various organs [7, 13]; however, their physiological roles in vivo remain to be fully clarified.
The three-dimensional (3D) structures of mammalian and yeast DAOs have been determined [14-17, 17a]. These studies revealed that the driving force of molecular interactions is the Coulombic force between an active site residue arginine guanidyl group of DAO and a carboxyl group of the substrate amino acid. A mutant, M213R of yeast DAO (PDB ID: 1C0P) [18] was prepared to analyze the freshness of food [19]. The stereo structure of sheep DAO was constructed from pig DAO (PDB ID: 1VE9), even the amino acid sequence of sheep DAO differs from that of 1VE9 by 8.5% to analyze the inhibition of the oxidation [20]. The structure of the docked complex of an enzyme, FAD, and a substrate was attempted to be optimized using the molecular mechanics 2 (MM2) calculation of the CAChe program. The tightness of the complex was determined as the energy value difference; however, the molecular interaction energy values were not quantitatively related to the enzyme reactivity. The tightness, but not the reactivity, was affected by the molecular size. Further calculation of the atomic partial charge (apc) using MOPAC-PM5 of CAChe program indicated the feasibility of the chemical reactions [1-3]. This approach was also applied to analyze enzyme reactivity of ELIS3-2 aromatic alcohol NADP+ oxidoreductase, even the amino acid identity was 33.4% from downloaded cinnamyl alcohol dehydrogenazse 1PIW [21].
Previously, the role of Ser-308 of mouse DDO was investigated through the use of site-specific mutational analysis of this residue [22]. In the report, alanine-, glycine-, and tyrosine-substitution mutants (S308A, S308G, and S308Y mutants, respectively) were prepared and their enzyme properties were characterized [22]. Here, the stereo structures of these mutants, in addition to a double-point mutant of mouse DDO (S308G-G309Y mutant), were constructed in silico and optimized using the MM2 calculation . Furthermore, apc of key atoms was calculated using a MOPAC-PM5 program, and then the reactivity of these mouse DDO mutants was analyzed.
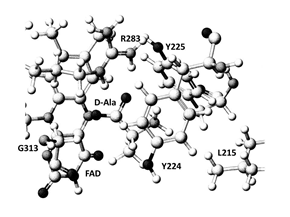
Figure1. Extracted stereo structures of hDAO; Black ball: oxygen, dark gray ball: nitrogen, large gray ball: carbon, small gray ball: hydrogen.
The stereo structure of human DAO (HDAO) was constructed using a stereo structure of pig DAO (PDB ID: 1VE9) [18] and a mammalian DAO sequence datum (NP001908) as performed for ELIS3-2 aromatic alcohol NADP+ oxidoreductase [21] by systematic replacement of amino acids (homology modeling) and optimized using MM2 calculation. The mammalian DAO indicated an amino acid identity of 85%, and shared a similarity of 90% of 340 amino acids [1], and the downloaded structure was corrected and optimized by a method [23]. Construction of above mutant was a little complicated compared to a thermostable mutant DAO from porcine kidney [24]; however, it was simpler than constructing ELIS3-2 oxidoreductase [21]. An original substrate D-alanine of the DAO was replaced to other amino acids by superimposing the carboxyl groups, and optimized by the MM2 calculation. The conformation of extracted structure is shown in Figure 1. The complexes demonstrated the lowest final structure energy values (data are not given) were selected, and the apc of key atoms was calculated using the MOPAC-PM5 program. The MOPAC calculation has limitation to handle numbers of atoms. Therefore, amino acid residues located within 3Å from the substrate were extracted with FAD, then locked the complexes, and calculated the apc. The apc values of a-carbon (αC) and a-nitrogen (αN) of substrate amino acids were corrected and are listed in Table 1 together with the specific activity (U mg-1 protein); one unit was defined as the production of 1mmol 2-oxo acid per min [25]. Furthermore, the feasibility of the constructed HDAO was analyzed by the comparison to four downloaded mammalian DAOs holding different molecular size substrates (PDB ID: 2du8 (benzoic acid), 3g3e (hydroxyquinoline-2(H)), 4qfc (3(4-hydroxy-6-[2-(7-hydroxy-2-oxo-4-phenyl-2h-chromen-6-yl)ethylpyridazin-3(2h)-one]), and 3znn (4H-thieno[3,2-b]pyrole-5-carboxylic acid) [18].
The stereo structure of mouse DDO was also constructed by homology modeling. These new stereo structures of DAO and DDO were optimized using the MM2 force-field parameters of version 6.1 CAChe MM2 (Fujitsu, Japan). The complexes of D-amino acid and the mutant were optimized. Amino acid residues existed within 3Å from D-amino acid were extracted with FAD, then locked the molecule, and optimized using PM5 program. The stereo structures are shown in Figure 2. Differences in the apc of key atoms and their relationships with enzyme activities were analyzed. Calculations were performed using a DCPIx86-based PC with an Intel Core™i7-2600 cpu 3.40 GHz. The minimum energy level was 10-7 kcal mol-1.
The expression plasmid for N-terminal His-tagged S308G mutant of mouse DDO (pRSET-His-mDASPO-S308G) has been described previously [22]. G309Y substitution was introduced into pRSET-His-mDASPO-308G by site-directed mutagenesis using the QuikChange ll Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clare, CA) to generate expression plasmid for the N-terminal His-tagged S308G-G309Y mutant (pRSET-His-mDASPO-S308G-G309Y). The following mutagenic oligonucleotides were used (mutated codons are underlined): 5’-ACG GCC ACG GGG GTT ACG GCA TCT CTG TGC ACT-3’ and 5’-AGT GCA CAG AGA TGC CGT AAC CCC CGT GGC CGT-3’. The mutation was confirmed by sequencing. A 1.1-kb NcoI-HindIII fragment of pRSET-His-mDASPO-S308G-G309Y was subcloned into pET-41a(+) (Novagen, Madison, WI) to generate expression plasmid for S308G-G309Y mutant of mouse DDO fused N-terminally with glutathione S-transferase-, His-, and S-tags (pET-GHS-mDASPO-S308G-G309Y). Expression in Escherichia coli, purification and enzymatic activity assays of the recombinant protein were carried out, as described previously [22]. E. coli BL21(DE3)pLysS cells were transformed with the expression plasmids and cultured at 37°C (with shaking) in Luria-Bertani medium containing 50 mg/mL kanamycin. Crude extracts were prepared from cells transformed with the expression plasmids as described previously [22].
All recombinant proteins were purified by affinity chromatography using a chelating column. Specifically, crude extracts (prepared as described above) were loaded onto a His GraviTrap column (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) equilibrated with 20 mM sodium phosphate buffer (pH 7.4) containing 0.5 M NaCl and 10 mM imidazole. The column was washed with the same buffer, and bound proteins were eluted using a stepwise gradient of 50–500 mM imidazole. Each fraction (2 mL) containing recombinant protein was mixed with FAD (at a final concentration of 50 mM) and dialyzed for 1 day at 4°C against 1 L of 10 mM sodium pyrophosphate buffer (pH 8.3) containing 2 mM EDTA, 5 mM 2-mercaptoethanol and 10% (v/v) glycerol. The buffer was changed once during dialysis. Dialyzed fractions were recovered and centrifuged at 20,000 × g for 10 min at 4°C to pellet any protein denatured during dialysis. Supernatants (purified enzyme preparations) were stored at -80°C until use.
The protein concentration of purified enzyme preparations was determined using the Bio-Rad Protein Assay Kit (Bio-Rad Laboratories, Hercules, CA) with bovine serum albumin as a standard. The oxidase activities of the purified enzymes were determined using a colorimetric method that measures 2-oxo acid production, as described previously [26]. Briefly, appropriate amounts (0.1–5.0 mg) of the purified enzymes were added to a reaction mixture containing 40 mM sodium pyrophosphate buffer (pH 8.3), 50 mM FAD, 33 ng/mL-1 catalase from Aspergillus niger (Sigma-Aldrich, St. Louis, MO) and 20 mM amino acids in a final volume of 150 mL. The reaction mixture was incubated at 37°C prior to addition of 10 mL of 100% (w/v) trichloroacetic acid to stop the reaction. The 2-oxo acid products were reacted with 2,4-dinitrophenylhydrazine and quantified by measuring the absorbance at 445 nm against a blank mixture lacking amino acids. To determine the Michaelis constant (Km) and maximal velocity (Vmax) values for D-aspartic acid and NMDA, these amino acids (at a final concentration of 1–40 mM) were used as substrates and the enzymatic reaction was performed under conditions in which the production of 2-oxo acids increased linearly with incubation time and exhibited standard Michaelis-Menten-type properties. The Km and Vmax values were calculated from Hanes-Woolf plots [27].
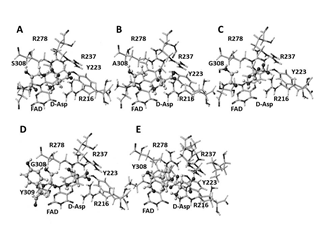
Figure2. Extracted stereo structures of DDO mutants optimized using molecular mechanics. A: DDO wild; B: S308A mutant; C: S308G mutant; D: S308G, G309Y mutant; E: S308Y mutant; Black ball: oxygen, dark gray ball: nitrogen, large gray ball: carbon, small gray ball: hydrogen; atomic size of the substrate D-aspartic acid is 20%, and others are 10%.
The center of electron transfer and oxidation reaction of amino acids is α-carbon (αC). The calculated apc values of αC of substrate amino acids are corrected and listed in Table 1 together with the specific activity (U mg-1 protein) [25].
Table 1. Enzyme activity and atomic partial charge (au) of α-carbon of D-amino acids
|
|
Enzyme |
apc of |
|
|||
|
Amino acid |
activity |
original |
apc of complex |
|||
|
|
U mg-1 protein |
form |
|
|||
|
Structure |
HDAO |
3znn |
3g3e |
2du8 |
||
|
D-Asparagine |
1.01 |
0.219 |
-0.177 |
-0.185 |
-0.172 |
-0.167 |
|
D-Glutamine |
ND |
-0.255 |
-0.221 |
-0.221 |
-0.225 |
-0.203 |
|
D-Alanine |
5.13 |
-0.249 |
-0.198 |
-0.194 |
-0.201 |
-0.195 |
|
D-Phenylalanine |
11.7 |
-0.256 |
-0.190 |
-0.147 |
-0.180 |
-0.162 |
|
D-Methionine |
0.83 |
-0.263 |
-0.216 |
-0.209 |
-0.216 |
-0.208 |
|
D-Valine |
2.00 |
-0.257 |
-0.216 |
-0.225 |
-0.218 |
-0.208 |
|
D-Leucine |
4.50 |
-0.247 |
-0.198 |
-0.180 |
-0.203 |
-0.180 |
|
D-Serine |
1.49 |
-0.260 |
-0.213 |
-0.215 |
-0.217 |
-0.219 |
|
D-Threonine |
0.81 |
-0.259 |
-0.222 |
-0.228 |
-0.223 |
-0.209 |
|
D-Tryptophan |
5.89 |
-0.264 |
-0.207 |
-0.208 |
-0.212 |
-0.196 |
|
D-Cysteine |
0.39 |
-0.235 |
-0.189 |
-0.170 |
-0.176 |
-0.188 |
|
D-Arginine |
0.30 |
-0.263 |
-0.231 |
-0.216 |
-0.218 |
-0.234 |
|
D-Lysine |
ND |
-0.250 |
-0.222 |
-0.199 |
-0.212 |
-0.202 |
|
D-Histidine |
0.74 |
-0.250 |
-0.219 |
-0.198 |
-0.214 |
-0.207 |
|
n |
14 |
14 |
14 |
14 |
14 |
|
|
r |
0.862 |
0.816 |
0.758 |
0.881 |
||
|
n (- D-Cysteine) |
13 |
13 |
13 |
13 |
13 |
|
|
r |
0.892 |
0.869 |
0.884 |
0.880 |
||
HDAO: human DAO as a mutant of pig DAO; 3znn, 3g3e, 2du8: human DAO
Previously, it was demonstrated that the enzyme reactivity (kcat/Km) of M213R mutant of yeast DAO [19] correlated well with apc of αC of substrate amino acids [2]. Therefore, the change of apc values (Δapc) were obtained by subtracting apc values of complexes with the enzyme from those of free amino acids. This approach was applied to study the specific activity of mammalian DAO. In this calculation, the specific activity values for D-glutamine and D-lysine were considered zero (U mg-1 protein).
For HDAO: U mg-1 protein = -0.28×10-2(Δapc) – 3.65×10-2, r = 0.862 (n = 14),
For 2du8: U mg-1 protein = -0.41×10-2(Δapc) – 4.34×10-2, r = 0.881 (n = 14),
For 3znn: U mg-1 protein = -0.50×10-2(Δapc) – 4.03×10-2, r = 0.863 (n = 14),
For 3g3e: U mg-1 protein = -0.26×10-2(Δapc) – 3.92×10-2, r = 0.758 (n = 14),
The larger the molecular size of a substrate, the easier is the docking (replacement). Therefore a downloaded HDAO (PDB ID: 4qfc) was used as standard structure; however, 4qfc lacks 10 substitute amino acids (Lys-338-to-Ser-340, Gly-299-to-Ser-301, and Pro-219-to-Gly-222); furthermore 6 amino acids that are supposed to be lysine appear as alanine. Therefore, the results used 4qfc were abandoned later. The conformation of D-cysteine and 3g3e complex was doubtful, and the result of D-cysteine was outliner. The correlation coefficient for 3g3e improved to 0.884 (n = 13) without D-cysteine. The correlation coefficient of other DAOs improved a little without D-cysteine. The slight difference in the 3g3e complex stereo structure may be due to the substrate (hydroxyquinoline). Other substrates connect with the guanidyl groups of Arg-283 via their carboxyl group. Therefore, the replacement after superimposed at carboxyl group demonstrated better correlation for their enzyme activity. The results are summarized in Table 1.
The correlation coefficient between the Δapc of αN of D-amino acids and the enzyme activity was poor. The apc values of ionized amino group of some D-amino acids were affected because of contacted Tyr-224 and/or Tyr-228, especially amino group of D-cysteine, D-threonine, and D-serine. The location of Tyr-224 was a little different among these protein structures. The correlation coefficient for HDAO, 3znn, 2du8, and 3g3e was 0.531, 0.465, 0.744, 0.569 (n = 14), respectively. The correlation coefficient improved a little without D-cysteine, and the values were 0.641, 0.436, 0.763, and 0.669 (n = 13), respectively. Above results demonstrated that enzyme activity can be quantitatively analyzed if the stereo structure of enzyme is available; however, we must carefully analyze the structure whether the structure has the right components and correct the structure of the co-enzyme.
Further analysis was carried out for the reactivity of several mutants of mouse DDO [22]. The stereo structure was constructed using a stereo structure of pig DAO (PDB ID: 1KIF [18]) and a mouse DDO sequence datum (NCBI: NM 027442.5). The mouse DDO indicated an amino acid identity of 43%, and shared a similarity of 56% of 189 amino acids. The apc of aC and aN of D-aspartic acid were correlated with the reactivity, Vmax/Km. The Vmax/Km and apc values are listed in Table 2. The reactivity for NMDA was also analyzed using the same approach, and the results are included in Table 2. The correlation for wild-type DDO reactivity and αC and αN of the substrate are shown as the following equations.
Table 2. Reactivity of several mutants of mouse DDO and apc of target atoms of D-aspartic acid and NMDA
|
|
For D-Aspartic acid For NMDA |
|||||
|
Mutant |
Vmax/Km |
apc αC |
apc αN |
Vmax/Km |
apc αC |
apc αN |
|
S308A |
3.146 |
-0.175 |
-0.017 |
1.432 |
-0.004 |
0.323 |
|
S308G |
3.479 |
-0.195 |
0.020 |
3.308 |
-0.054 |
0.342 |
|
S308S (Wild type) |
2.874 |
-0.174 |
-0.034 |
0.468 |
0.002 |
0.317 |
|
S308G-G309Y |
3.241 |
-0.196 |
0.015 |
2.204 |
-0.060 |
0.355 |
|
S308Y |
2.606 |
-0.154 |
-0.023 |
0.363 |
0.020 |
0.326 |
Unit: apc (au)
For D-aspartic acid:
Vmax/Km = -42.849 (Δapc of αC) + 1.472, r = 0.849 (n= 5)
Vmax/Km = 32.203 (Δapc of αN) – 7.903, r = 0.876 (n = 5)
For NMDA:
Vmax/Km = -31.245 (Δapc of αC) + 0.955, r = 0.902 (n= 5)
Vmax/Km = 58.833 (Δapc of αN) – 18.013, r = 0.741 (n = 5)
The calculates apc values were related to Vmax/Km. S308G demonstrated the strong reactivity for both D-aspartic acid and NMDA, and S308Y did the weak reactivity, however, G309Y did not affect the reaction.
Previously, we performed conformation analysis of HDAO complex with several active site-directed inhibitors [28, 29], but this analysis did not quantitatively demonstrate the enzyme reactivity (inhibitory activity). On the other hand, the reactivity can be quantitatively described by electron localization of key atoms as used in organic synthesis chemistry. Here, we demonstrated that the apc of key atoms can support the oxidation reaction of DAO and DDO. G183 is located near phosphate of FAD and is far from the reaction center; however, G183R [30] may affect the conformation of the reaction chamber as FMN did [1]. Further study is necessary to make clear the conformation, as also obtaining useful insight into the biological significant of human DDO based on the biological study for rat and human DDO [31]. The method described herein may contribute to basic enzymology and application to the development of useful enzyme inhibitors.
The stereo structures of mutants of DAO and DDO were constructed by homology modeling in silico and optimized using the MM2 calculation. Furthermore, apc of key atoms was calculated using a MOPAC-PM5 program. The oxidation reactivity for amino acids was correlated well with apc of C of substrate D-amino acids. Even conformation analysis of mammalian DAO complex with several active site-directed inhibitors did not quantitatively demonstrate the enzyme reactivity. However, the reactivity can be quantitatively described by electron localization of key atoms as used in teaching organic synthesis chemistry. Above results demonstrated that the apc of key atoms can support the oxidation reaction of DAO and DDO.
ABBREVIATIONS
DAO, D-amino acid oxidase; DDO, D-aspartate oxidase; FAD, flavin adenine dinucleotide; NMDA, N-methyl-D-aspartate; MM2, molecular mechanics 2; apc, atomic partial charge; au, atomic unit
Hanai, T. Quantitative in silico analysis of molecular recognition and reactivity of D-amino acid oxidase. Intern. Electr. J. Mol. Design, 2006, 5: p. 247-259.
Hanai, T. Quantitative in silico analysis of enzyme reactions: comparison of D-amino acid oxidase and monoamine oxidase. Am. Biotechnol. Lab. 2007, 25, p. 8-13.
Hanai, T. Quantitative in silico analysis of D-amino acid oxidase reactivity and inhibition, Current Bioactive Compounds, 2017, 13, p. 312-317. DOI: 10.2174/1573407212666161014133246.
View ArticleNithya, S; Shanmugarajam, T.S. D-Amino acid oxidase: A review. Asian J. Pharm. Clin. Res., 2016, 9, p. 33-36.
Panthis, S.; Chung, H.-J.; Jung, J.; Jeong, N.Y. Physiological importance of hydrogen sulfite: Emerging potent neuroprotector and neuromodulator. Oxid. Med. Cell. Longev., 2016, 9049781.
Kimura, H. Signalino molecules: Hydrogen sulfide and poly sulfide. Antioxid. Redox. Signal., 2016, 22, p. 362-375. PMid:24800864 PMCid:PMC4307032
View Article PubMed/NCBIOhide, H.; Miyoshi, Y.; Maruyama, R.; Hamase, K.; Konno, R. D-Amino acid metabolism in mammals: biosynthesis, degradation and analytical aspects of the metabolic study. J. Chromatogr. B, 2011, 879, p. 3162-3168. PMid:21757409
View Article PubMed/NCBINishikawa, T. Analysis of free D-serine in mammals and its biological relevance. J. Chromatogr. B, 2011, 879, p. 3169-3183. PMid:21992750
View Article PubMed/NCBIWolosker, H. NMDA receptor regulation by D-serine: new findings and perspectives. Mol. Neurobiol. 2007, 36, p. 152-164. PMid:17952659
View Article PubMed/NCBIKakegawa, W.; Miyoshi, Y.; Hamase, K.; Matsuda, S.; Matsuda, K.; Kohda, K.; Emi, K.; Motohashi, J.; Konno, R.; Zaitsu, K.; Yuzaki, M. D-Serine regulates cerebellar LTD and motor coordination through δ2 glutamate receptor. Nature Neurosci. 2011, 14, p. 603-611. PMid:21460832
View Article PubMed/NCBIDi Fiore, M.M.; Santillo, A.; Baccari, G.C. Current knowledge of D-aspartate in glandular tissues, Amino Acids, 2014, 46, p. 1805-1818. PMid:24839076
View Article PubMed/NCBIKatane, M.; Homma, H. D-Aspartate – an important bioactive substance in mammals: a review from an analytical and bioanalytical point of view. J. Chromatogr. B. 2011, 879, p. 3108-3121. PMid:21524944
View Article PubMed/NCBIKatane, M.; Homma, H. D-Aspartate oxidase: the sole catabolic enzyme acting on free D-aspartate in mammals. Chem. Biodivers, 2010, 7, p. 1435-1449. PMid:20564562
View Article PubMed/NCBIMattevi, A.; Vanoni, M.A.; Todone, F.; Rissi, M.; Teplyakov, A.; Coda, A. Bolognsi, M., Curti, B., Crystal structure of D-amino acid oxidase; a case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl. Acad. Sci. U.S.A. 1996, 93, p. 7496-7501. PMid:8755502
View Article PubMed/NCBIMizutani, H.; Miyayama, I.; Hitotsu, K.; Nishina, Y.; Shiga, K. Setoyama, C.; Miura, R. Three-dimensional structure of porcine kidney D-amino acid oxidase at 3.0Å resolution. J. Biochem. 1996, 120, p. 14-17. PMid:8864836
View Article PubMed/NCBIUmhau, S.; Pollegioni, L.; Molla, G.; Diederichs, K.; Welte, W.; Pilone, M.S.; Ghisla, S. The X-ray structure of D-amino acid oxidase at very high resolution identifies the chemical mechanism of flavin-dependent substrate dehydrogenation. Proc. Natl. Acad. Sci. U.S.A. 2000, 97, p. 12463-12468. PMid:11070076 PMCid:PMC18786
View Article PubMed/NCBIKawazoe, T.; Tsuge, H.; Pilone, M.S.; Fukui, K. Crystal structure of human D-amino acid oxidase: context-dependent variability of the backbone conformation of the VAAGL hydrophobic stretch-located at the si-face of the flavin ring. Protein Sci. 2006, 15, p. 2708-2717. PMid:17088322 PMCid:PMC2242440
a. Kawazoe, T., Tsuge, H.; Imagawa, T.; Aki, K.; Kuramitsu, S.; Fukui, K. Structural basis of D-DOPA oxidation by D-amino acid oxidase: Alternative pathway for dopamine biosynthesis, Biochem. Biophys. Res., 2007, 355, p. 385-391. https://doi.org/10.1016/j.bbrc.2007.01.181 PMid:17303072
RCSB, PDB protein data bank, www.rcsb.org/pdb/.
Sacchi, S.; Lorenzi, G.; Molla, M.S.; Pilone, C.; Rossetti, L.; Pollegioni, L. Engineering the substrate specificity of D-amino acid oxidase. J. Biol. Chem. 2002, 277, p. 510-516. PMid:12021281
View Article PubMed/NCBIDe Marche, W.J.; Johnston, G.A.R. The oxidation of glycine by D-amino acid oxidase in extracts of mammalian central nervous tissue. J. Neurochem., 1969, 16, p. 355-361.
View ArticleHanai, T. Quantitative in silico analysis of alcohol dehydrogenase reactivity, J. Drug Design & Research, 2015, 2, 1006-1012.
Katane, M.; Hanai, T.; Furuchi, T.; Sekine, M.; Hommam, H. Hyperactive mutants of mouse D-aspartate oxidase: mutagenesis of the active site residue serine 308. Amino Acids, 2008, 35, p. 75-82. PMid:18235994
View Article PubMed/NCBIHanai, T. Basic properties of a molecular mechanics program and the generation of unknown stereo structures of proteins for quantitative analysis of enzyme reactions, In: Watkins, P. (Ed.), Molecular Mechanics and Modeling. Nova Science: New York, 2015, Chapter 2, p. 25-48.
Bakke, M.; Setoyama, C.; Miura, R.; Kajiyama, M. Thermostabilization of porcine kidney D-amino acid oxidase by a single amino acid substitution, Biotechnol. Bioeng., 2006, 93, p. 1023-1027. PMid:16245349
View Article PubMed/NCBIKatane, M.; Saitoh, Y.; Maeda, K.; Hanai, T.; Sekine, M.; Furuchi, T.; Homma, H. Role of the active site residues arginine-216 and arginine-237 in the substrate specificity of mammalian D-aspartate oxidase. Amino Acids, 2011, 40, p. 467-476. PMid:20567862
View Article PubMed/NCBIKatane, M.; Seida, Y.; Sekine, M.; Furuchi, T., Homma, H. Caenorhabditis elegans has two genes encoding functional D-aspartate oxidase. FEBS J. 2007, 274, p. 137-149. PMid:17140416
View Article PubMed/NCBIHanes, CS.; Studies on plant amylases: the effect of starch concentration upon the velocity of hydrolysis by the amylase of germinated barley, Biochem. J. 1932, 26, p. 1406-1421. PMid:16744959 PMCid:PMC1261052
View Article PubMed/NCBIKatane, M.; Osaka, N.; Matsuda, S.; Maeda, K.; Kawata, T.; Saitoh, Y.; Sekine, M.; Furuchi, T.; Doi, I.; Hirono, S.; Homma, H. Identification of novel D-amino acid oxidase inhibitors by in silico screening and their functional characterization in vitro. J. Med. Chem. 2013, 56, p. 1894-1907. PMid:23391306
View Article PubMed/NCBIKatane, M.; Matsuda, S.; Saitoh, Y.; Sekine, M.; Furuchi, T.; Koyama, N.; Nakagome, I.; Tomoda, H.; Hirono, S.; Homma, H. The activiral drug acyclovir is a slow-binding inhibitor of D-amino acid oxidase. Biochemistry 2013, 52, p. 5665-5674. PMid:23859606
View Article PubMed/NCBIMurtas, G.; Caldinell, L.; Cappelletti, P., Sacchi, S.; Pollegioni, L. Human D-amino acid oxidase: The inactive G183R., Biochim. Biophys. Acta Proteins and Proteomics, 2018, 1866, p. 822-830.
Katane, M.; Kuwahara, H.; Nakayama, K.; Saitoh, Y.; Miyamoto, T.; Sekine, M.; Homma, H. Rat D-aspartate oxidase is more similar to the human enzyme than the mouse enzyme, Biochim. Biophys. Acta-Proteins and Proteomics, 2018, 1866, p. 806-812.
