Xinhong Guo, Email: gxh@hnu.edu.cn
Jiazhuo Ye, Email: yjz@hnu.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
Xinhong Guo, Email: gxh@hnu.edu.cn
Jiazhuo Ye, Email: yjz@hnu.edu.cn
Meiling Li1,†, Xiaoxia Lin1,†, Xiutao Lu1,†, Shucan Liu1, Huili Li1, Cheng Zhang1, Yan Lu1, Xinhong Guo1*, Jiazhuo Ye1*
1 College of Life Sciences, Hunan University, Changsha 410082, P.R. China
2 College of Electrical and Information Engineering, Hunan University, Changsha 410082, P.R. China
†These authors contributed equally to this work
xinhong guo, LecRKIII.1 and LecRKIII.2 formed homodimers to play physiological functions in Arabidopsis thaliana(2018)SDRP Journal of Plant Science 2(2)p:115-123
Abstract:Receptor-like kinases (RLKs), an important family that plant cell sense and transmit extracellular signals, are fundamental to plant cell life and play critical roles in various biological processes, such as response to abiotic stress, development and hormonal response. Among RLKs, the lectin receptor-like kinases (LecRLKs) possess a characteristic extracellular carbohydrate-binding lectin domain and play important roles in plant immune, stress and hormone responses. However, little is known about the genes LecRKIII.1 and LecRKIII.2 of LecRLK family up to now. Here, we founded that LecRKIII.1 and LecRKIII.2 were localized in plasma membrane and played the physiological roles by forming a homodimer. Real-time fluorescence quantitative PCR analysis of the expression level of LecRKIII.1 and LecRKIII.2 in response to exogenous ABA, MeJA, SA, NaCl, mannitol and dehydration suggested that LecRKIII.1 and LecRKIII.2 may play important roles in response to various stresses. It will lay a good foundation for further studies on physiological functions of LecRKIII.1 and LecRKIII.2.
Key words :LecRLK, Yeast two hybrid, BiFC, cDNA library, qRT-PCR
Compared with mobile animals, sessile plants evolved developmental plasticity as an important strategy to adapt to different environmental conditions [1]. In other words, plants must depend on a complicated signaling system in response to external stimuli. Arabidopsis thaliana receptor-like kinases (RLKs) are widespread proteins family in all kingdoms of plant life and are represented by 610 genes in Arabidopsis genome [2]. RLKs contain an extracellular receptor domain, a single transmembrane domain and an intracellular kinase domain in plant [3]. The extracellular receptor domain specifically recognizes and binds extracellular signal molecules to alter the conformation of the intracellular kinase domain by homologous or heterodimerization, and then activate intracellular serine /threonine kinase domain and phosphorylate downstream proteins, which will recongnize and transfer the extracellular signal into the cell [3]. Generally, in plants, RLKs are the ideal candidates to percept extracellular stimuli and further initiate signaling pathways [4,5,6]. RLKs are generally divided into 6 categories: S RLKs, Ieucine-rich repeats RLK(LRRs), Chitinase-related RLKs (CR4), WAK-1ikeRLKs, TNFR.1ike RLKs, PR5-RLKs, LecRLKs, based on the difference of the RLKs extracellular receptor domain.
LecRLKs, which are characterized by an N-terminal lectin domain and are represented by 75 members in Arabidopsis [7,8,9]. On the basis of the extracellular lectin domains’difference, the LecRLKs family could be further subdivided into three categories: L-type, G-type and C-type LecRLKs [10]. Here, LecRKlll.1 and LecRKlll.2 belong to L-type LecRKs, which are ubiquitous in plants and have been identified in avarieties of plant species, such as cotton and rice [11,12]. Thus far, with the continuous study of LecRLKs family, they are thought to be play important roles in microbial identification and response [13]. For example, plants could respond to pathogens by activation of plasma membrane-localized receptor-like kinases (RLKs) that function as pattern recognition receptors (PRRs) to initiate defense [14,15,16]. Recently, it has been demonstrated that LecRK-VI.2 (At5g01540) recognized the pathogen-related molecular component β-aminobutyric acid induce stomatal closure to enhance the resistance of plants to pathogens such as pseudomonas syringae [17,18]. A. thaliana LecRK-I.8 played a great role in defense triggered by eggderived elicitors of the cabbage butterfly Pieris brassicae [19]. In addition, few L-type LecRKs are involved in plant other biotic and abiotic stress. The LecRLKs responsing to wounding were reported for Arabidopsis LecRK-V.5 and Populus nigra PnLPK genes [20,21,22]. Arabidopsis LecRLKs family A4 subfamily member gene LecRK-b2 participated in plant hormone abscisic acid signal transduction pathway [23].
In conclusion, The Arabidopsis L-type LecRLKs family genes have important research significance. However, the function of genes LecRLKIII.1 and LecRLKIII.2 and function mechanism of the LecRLKs family have not been studied. In this study, we employed Yeast two-hybrid and BiFC experiments for confirming that LecRKIII.1 can interact with itself to form homodimers and LecRKIII.2 also have similar function. In addition, our datas suggested that the genes LecRKIII.1 and LecRKIII.2 may play important roles in response to multiple stresses.
Plant materials and growth conditions
Arabidopsis ecotype Columbia-0 (Col-0) was used as the materials. Seeds of the wild type Col-0 were surface-sterilized in 75% alcohol solution and 20% sodium hypochlorite for 8 min and then washed four times in sterile distilled water. They were plated on 0.8 % (w/v) solid agar MS medium at 4 °C for 3 d synchronization to promote their germination rate. After7 d, the green seedlings were transferred to soil and continually grown at 22 °C under long-day conditions (16 h light and 8 h dark) until the last siliques were dry, and seeds were harvested and stored as described [24].
For treatment analyses, seven-day-old wild-type seedlings were selected as the materials. Firstly, the seedlings were gently pulled out from the MS plate. After soaking, the seedlings were treated with 100 mM ABA, 100 mM MeJA, 100 mM SA, 100 mM mannitol and 100 mM NaCl in the MS liquid medium, respectively. Materials were collected at several time points. All samples were frozen shortly in liquid nitrogen to keep them fresh for the next experiments.
RNA isolation and qRT-PCR Analysis
Total RNA was extracted from the frozen samples of Arabidopsis by using a total RNA Purifcation System kit (Invitrogen) and cDNA was synthesized using a PrimeScriptTM RT reagent kit (Takara). For quantitative real-time PCR analysis, we carried out three biological replicates to guarantee precise statistical results. RNA accumulation of genes in each sample was normalized to that of Actin2 gene. The following amplifcation procedure was used: 94 °C for 10min, followed by 40 amplifcation cycles of 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Finally, we analyzed the qPCR results using software MXPro. The primers used in the qRT-PCR reactions are presented in Table 1.
Table 1. Primer sequences were used in quantitative RT-PCR.
|
Gene |
Primer Name |
Sequence 5’-3’ |
|
LecRKIII.1 |
At2g29220QF |
GACCGACCAGTTCTCACC |
|
At2g29220QR |
GTGTTACCTTTCCCGTGT |
|
|
LecRKIII.2 |
At2g29250QF |
ACGACCTACCAGTTCTCA |
|
At2g29250QG |
GTGTCCCGTAGTGCTGTG |
Yeast two hybrid and BiFC
The pGBKT7 and pGADT7 vectors and the yeast strain AH109 were used for Yeast two hybrid assays. The C-terminal sequences and N-terminal sequences of LecRKIII.1 and LecRKIII.2 were both cloned into pGBKT7 and pGADT7. The recombinant constructs and empty vectors were transformed into yeast strain AH109 in pairs. Transformants spotted on plates of selective medium without Leu and Trp (SD-Leu-Trp) indicating that two vectors pGADT7 and PGBKT7 that expressed LecRKIII.1 and LecRKIII.2 of the N-terminal and C-terminal fusion proteins were successfully transferred, respectively. Next, the cells were plated on SD medium lacking leucine, tryptophan and histidine, (SD-Leu-Trp-His) supplemented with 3-amino-1,2,4-triazole for screening. All vectors and primers used in Yeast two hybrid and BiFC assays are presented in Table 2 and Table 3.
Table 2. C/N-terminal primers of LecRKIII.1 and LecRKlII.2 were used in Yeast two hybrids.
|
Primer Name |
Sequence 5’-3’ |
RestrictionEnzymeCutting Site
g Site Cutting Site |
|
220C-pGBKT7 F
|
CGGAATTCTGGTTCTTACGTAGAGACAAAATATTC
|
EcoR I EcoR I |
|
220C-pGBKT7R
|
CGGGATCCTTAGACTCCATAGAAGAGGTAGTCATC
|
BamH I
|
|
250C-pGBKT7 F
|
CGGAATTCTGCTTTTTTCGTAGAACGGAAAACTTC
|
EcoR I EcoR I |
|
250C-pGBKT7 R
|
CGGGATCCTTAGACGACGTAGTCATCATGAGGGAG
|
BamH I
|
|
220C-pGADT7 F
|
CGGAATTCTGGTTCTACGTAGAGACAAAATATTC
|
EcoR I EcoR I |
|
220C-pGADT7 R
|
CGGGATCCTTAGACTCCATAGAAGAGGTAGTCATC
|
BamH I
|
|
250C-pGADT7 F
|
CGGAATTCTGCTTTTTTCGTAGAACGGAAAACTTC
|
EcoR I |
|
250C-pGADT7 R
|
CGGGATCCTTAGACGACGTAGTCATCATGAGGGAG
|
BamH I
|
|
250N-pGBKT7 F
|
GGAATTCCATATGCAACAACAAACCAAATTTCTTACC
|
Nde I
|
|
250N-pGBKT7 R
|
CGCGGATCCCATCGTGTTACTCGAATTTTTCAT
|
BamH I
|
|
220N-pGBKT7 F
|
GGAATTCCATATGCAACGACAGACCAAGTTTCTTAACC
|
Nde I
|
|
220N-pGBKT7 R
|
CGCGGATCCCATCGTGTTGTTCGACTTCTTCTC
|
BamH I
|
|
250N-pGADT7 F
|
GGAATTCCATATGCAACAACAAACCAAATTTCTTAACC
|
Nde I
|
|
250N-pGADT7R RR
|
CGCGGATCCATCGTGTTACTCGAATTTTTCAT
|
BamH I
|
|
220N-pGADT7 F
|
GGAATTCCATATGCAACGAGAGACCAAGTTTCTTAACC
|
Nde I
|
|
220N-pGADT7R RRR
|
CGCGGATCCCATCGTGTTGTTCGACTTCTTCTC
|
BamH I
|
Table 3. All primers were used in BiFC assays.
|
Primer Name |
Sequence 5’-3’ |
Restriction Enzyme Cutting Site |
|
250-3308-F |
GGGGTACCTCATGGCTAATACCTATAAATCGATAGCTG |
Kpn I |
|
250-3308-R |
TCCCCCGGGGACGACGTAGTCATCATGAGGGAG |
Sma I |
|
250-3349-F |
GGGGTACCTCATGGCTAATACCTATAAATCGATAGCTG |
Kpn I |
|
250-3349-R |
TCCCCCGGGGACGACGTAGTCATCATGAGGGAG |
Sma I |
|
220-3308-F |
GGGGTACCTCATGATTACCTTCAAATCTATAGCTCTAAC |
Kpn I |
|
220-3308-R |
TCCCCCGGGGACTCCATAGAAGAGGTAGTCATCAT |
Sma I |
|
220-3349-F |
GGGGTACCTCATGATTACCTTCAAATCTATAGCTCTAAC |
Kpn I |
|
220-3349-R |
TCCCCCGGGGACTCCATAGAAGAGGTAGTCATCAT |
Sma I |
Subcellular Localization
To determine the subcellular localization of LecRKIII.1 and LecRKIII.2 in Arabidopsis thaliana cells, the coding sequence of LecRKIII.1 and LecRKIII.2 were amplified by PCR using the primer pair LecRKIII.1/LecRKIII.2 (F/R) (Table 4) and then cloned into the vector pEZS-NL to generate pEZS-NL-LecRKIII.1/LecRKIII.2. Then, recombinant 35Spro-GFP-LecRKIII.1 and 35Spro-GFP-LecRKIII.2 were introduced into Arabidopsis protoplasts, respectively. The transient GFP fluorescence in A. thaliana leaf cells was observed under a Leica SPE confocal microscope as described previously [25,26].
Table 4. Primer sequences used in the coding sequence were amplification of LecRKIII.1 and LecRKlII.2 in subcellular localization.
|
Primer Name |
Sequence 5’-3’ |
Restriction Enzyme Cutting Site |
|
PEZS-250F |
GGGGTACCTATGGCTAATACCTATAAATCGATAGCTG
|
Kpn I Kpn I |
|
PEZS-250R |
CGGGATCCGGGACGACGTAGTCATCATGAGGGAG
|
Bam HI |
|
PEZS-220F |
GGGGTACCTATGATTACCTTCAAATCTATAGCTCTAAC
|
Kpn I |
|
PEZS-220R |
CGGGATCCGGGACTCCATAGAAGAGGTAGTCATCAT
|
Bam HI |
LecRKIII.1 and LecRKIII.2 were localized on plasma membrane
We predicted that 97.5% of LecRKIII.1 and 97.1% of LecRKIII.2 are located on the plasma membrane, respectively by softberry website (http://linux1.softberry.com/) . To further verify the authenticity of the prediction, subcellular localizations of LecRKIII.1 and LecRKIII.2 have been completed. We expressed GFP-LecRKIII.1 and GFP-LecRKIII.2 fusion protein under the control of the 35S promoter in wild-type plants, respectively. Transient expression of GFP-LecRKIII.1 or GFP-LecRKIII.2 in Arabidopsis leaf protoplasts showed that both LecRKIII.1 and LecRKIII.2 were localized in the plasma membrane (Fig. 1), this is consistent with the previous bioinformatics analysis results.
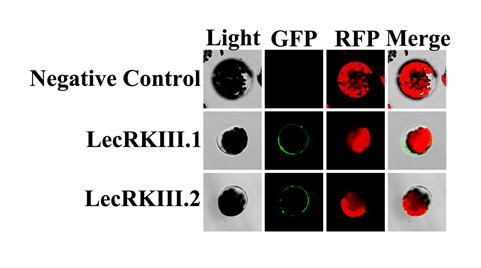
Fig. 1 LecRKIII.1 and LecRKlII.2 Localize to plasma membrane. From top to bottom, Negative control, fusion protein GFP- LecRKIII. 1+LecRKIII. 1, fusion protein GFP-LecRKIII. 1+LecRKIII. 2. From left to right, the light, fluorescence of GFP, fluorescence of RFP and merging of GFP and RFP.
LecRKIII.1 and LecRKIII.2 can interact with themselves in the form of homodimer
Here, we have already proved that LecRKIII.1 and LecRKIII.2 were localized on plasma membrane, and LecRKIII.1 and LecRKIII.2 were predicted to contain a N-terminal extracellular domain (amino acids residues 24 to 303) with glycosylation site and ATP Binding site, a C-terminal cytoplasmic domain (residues 324 to the final amino acid) and single transmembrane proteins through the UniProt Web site (http://www.uniprot.org/). These are common with the existing receptor kinases, its extracellular glycosylation site and ATP Binding site can combine with extracellular signal molecules and have kinase domain, separately. Moreover, receptor kinases usually conduct biological functions in the form of dimers. To clarify whether LecRKIII.1 and LecRKIII.2 play a role through forming dimers, we conducted yeast two hybrid. As shown in Fig. 2A, only the yeast cell that contained C-terminal domain of LecRKIII.1/2 and C-terminal domain of LecRKIII.1/2 could grow appropriately on SD/-Trp−Leu−His screening medium, suggesting that the C-terminal domain of LecRKIII.1/2 can able to interact with themselves to form homodimers. To further explore this result, we conducted biomolecular fluorescence complementation (BiFC), no green fluorescence was found in the negative control, whereas when the vectors that expressed fusion protein LecRKIII.1 or LecRKIII.2 were transferred into Arabidopsis protoplasts, green fluorescence was observed ( Fig. 2C) . The results are consistent with those obtained above in yeast two hybrid experiments. These findings demonstrated that proteins LecRKIII.1 and LecRKIII.2 were able to interact with themselves to form homodimers.
LecRKIII.1and LecRKIII.2 are predicted to have 85% gene sequence and 82% protein sequence similarity, suggesting these two genes have high homology (https://www.arabidopsis.org/Blast/). We have proved that LecRKIII.1 can interact with itself to form homodimers and LecRKIII.2 also has similar function. To further ask whether LecRKIII.1 and LecRKIII.2 can also interact with each other, we performed yeast two-hybrid assays again. The results showed that both N-terminal and C-terminal of LecRKIII.1 were not able to interact with N-terminal and C-terminal of LecRKIII.2 (Fig. 2B), demonstrating that LecRKIII.1 and LecRKIII.2 didn`t form a heterodimer.
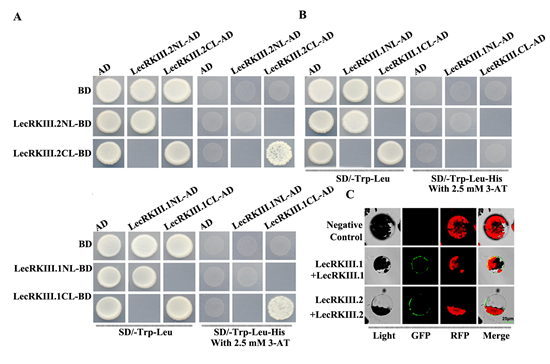
Fig. 2 Protein protein interact analysis of LecRKIII.1 and LecRKIII.2 by Y2H and BiFC.
(A) LecRKIII.1 and LecRKIII.2 play a role through forming dimers using yeast two-hybrid screening. AD, activationdomain; BD, DNAbindingdomain. Transformed yeasts were spotted on control medium(SD-Leu-Trp) or selective medium (SD-Leu-Trp-His ) supplemented with 3-amino-1,2,4-triazole for screening. The empty vectors were used as controls.
(B) Testing of interactions between LecRKIII.1 and LecRKIII.2.
(C) BIFC assays proved that LecRKIII.1 and LecRKIII.2 were able to interact with themselves to form homodimers.
Expression of the LecRKIII.1 and LecRKIII.2 gene in response to various stresses
To understand and predict results of LecRKIII.1 and LecRKIII.2 in Arabidopsis in more detail, we performed RT-PCR analysis at several time points to explore their expression patterns under various abiotic stress treatments. Seven-day-old seedlings were treated with ABA, MeJA, SA, mannitol, NaCl and dehydration stress, respectively. The expressions of LecRKIII.1 and LecRKIII.2 in response to exogenous ABA, MeJA, SA, NaCl, mannitol and dehydration were shown in Fig. 3. The expression of LecRKIII.1 and LecRKIII.2 decreased when treated with ABA, whereas the expression of LecRKIII.2 after 8h treatment fluctuated, which may be related with its negative feedback pathway (Fig. 3A). The expression of LecRKIII.1 and LecRKIII.2 were significantly induced by dehydration stress, peaking at 6 h (Fig. 3D), by NaCl at 8h (Fig. 3F), by MeJA at 6h and 12h (Fig. 3B) and by mannitol at 9 and 6h for LecRKIII.1 and LecRKIII.2, respectively (Fig. 3E). Interestingly, there are no regular changes when treated with SA, showing that LecRKIII.1 and LecRKIII.2 may don’t participate in SA signal path (Fig. 3C). Collectively, the outcome strongly suggested that LecRKIII.1 and LecRKIII.2 might be involved in various stresses, but not in the SA stress response signaling pathways.
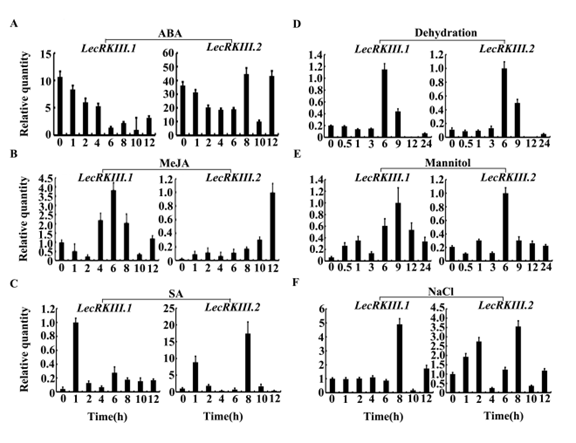
Fig. 3 Real-time fluorescence quantitative PCR analysis of the expression level of LecRKIII.1 and LecRKIII.2 in response to exogenous ABA (A), MeJA (B), SA (C), dehydration stress (D), mannitol (E) and NaCl (F) respectively. Actin-2 was used as control.
In this study, we identified that LecRKIII.1 and LecRKIII.2 have typical L-type LecRLK structure-an extracellular soybean lectin domain, a single transmembrane region and a cytoplasmic serine/threonine kinase domain by bioinformatics analysis. Sequence alligment demonstrated that LecRKIII.1 and LecRKIII.2 have a very high homology. Here, we proposed a function mechanism for LecRKIII.1 and LecRKIII.2 in which LecRKIII.1/2function as homogous dimers. However, the yeast two hybrid confirmed that LecRKIII.1 and LecRKIII.2 didn`t interact with each other to form a heterodimer.
Previous studies have shown that RLKs family function as homodimers, heterodimers or oligomers, but the function mechanisms of LecRKIII.1 and LecRKIII.2 were not investigated [27,28,29,30,31,32]. Our study demonstrated that LecRKIII.1 and LecRKIII.2 could interact with themselves to form homodimers. Although there were 85% gene sequence and 82% protein sequence similarity between LecRKIII.1 and LecRKIII.2, they didn`t interact with each other to form a heterodimer, we speculated that LecRKIII.1 may function in a redundant manner with LecRKIII.2, but they function independently.
Plants are often exposed to various adverse environment that greatly is harmful to growth. To adapt to all kinds of complex external conditions, they have formed a set of fine strategies for survival. One of the key measure is that RLKs transmit extracellular signals to intracellular. Therefore, RLKs are crucial in regulating plant reactions to biotic and abiotic stress [33,34], and previous studies have also proved this view [35,36]. However, no study showed that LecRKIII.1 and LecRKIII.2 are related to hormone and stress response in details. And largely, abscisic acid (ABA), salicylic acid (SA), jasmonic acid (JA), MeJA, NaCl and mannitol are key regulations of plant growth and development. In our conclusion, we founded that there were certain regular expression level changes except SA under different treatment. We found that LecRKIII.1 and LecRKIII.2 have the same expression pattern under dehydration, NaCl and mannitol. The expression level of LecRKIII.1 and LecRKIII.2 peaking at 6, 8 or 9 h. However, they presented opposite expression mode when treated with MeJA. The expression level of LecRKIII.1 reached maximum at 6 hours and then declined, but the expression of LecRKIII.2 was activated and increased after the expression declining of LecRK.III.1. These results suggested that LecRKIII.1 and LecRKIII.2 may play important roles in response to multiple stresses.
In summary, the present research provides a significant work mechanism that LecRKIII.1 and LecRKIII.2 function as homogous dimmers. It is beneficial for deep research of LecRKs family. The expressions of LecRKIII.1 and LecRKIII.2 were induced with the above-mentioned different ways of treatment, suggesting that these two genes might modulate these various signalling pathways and ultimately enhance Arabidopsis thaliana’s tolerance in response to different intrinsic and environmental signals.
This research was supported by grants from National Key Research and Development Program (2017YFF0210301), Key Research & Development Project of Hunan Provincial Department of Science and Technology (2016WK2003 and 2017SK218), National Natural Science Foundation of China (31540064, 31071076 and 30871325), the National Key Laboratory of Plant Molecular Genetics (2015), and Hunan Provincial Innovation Foundation for Postgraduate (CX2017B145, CX2015B073, and CX2016B097).
