Francesco Villani
E-mail address: villani@unina.it
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 6 ISSUE: 1
Page No: 299-312
Francesco Villani
E-mail address: villani@unina.it
Marina Gielloa, Antonietta La Storiaa, Paola Zinnob,Barbara Guantariob, Francesco Villania*
a Department of Agricultural Science, Division of Microbiology, University of Naples Federico II, Via Università100, 80055, Portici (NA), Italy.
b Research Centre for Food and Nutrition, CREA (Council for Agricultural Research and Economics),Via Ardeatina 546, 00178 Rome, Italy.
Marina Giello, Antonietta La Storia, Paola Zinno,Barbara Guantario, Francesco Villani, Autochthonous Lactobacillus paracasei subsp. paracasei strains isolated from Caciocavallo cheese: identification and in-vitro investigation on potential probiotic and functional properties (2021) Journal of Food Science & Technology 6(1) pp:299-312
Background: In recent years consumers, due to the growing interest for functional foods, have put particularly attention on products enriched with probiotic lactic acid bacteria for their ability to give human benefit by their assumption. Microorganisms to be considered as probiotic should be able to survive to gastrointestinal juices and colonize the intestinal tract to provide benefits to human health.
The aim of the study consisted of the in vitro investigation of novel LAB strains, isolated during cheesemaking and ripening of Caciocavallo cheese, and on their probiotic potential for food application.
Methods: In order to select potential probiotic bacteria, LAB isolates were preliminarily screened for their survival to simulated gastro-intestinal transitand assayed for other activities of probiotic and functional interest. Thanks to promising attitude as potential probiotics, four strains were tested to evaluate their adhesion ability on Caco-2 cell lines used as intestinal cell model.
Microbiological data were analysed by one-way analysis of variance (ANOVA). Comparisons of means were carried out using post-hoc Tukey’s test. For vitro adhesion experiments, statistical significance was evaluated by Kruskal-Wallis test for equal medians.
Results: Twenty-five LAB, presumptively identified as Lactobacillus spp., were isolated from cheese during 60 days of ripening and screened for their survival to simulated gastrointestinal transit. Sixteen isolates that showed a survival rate ≥85% were identifiedas 10 different profiles of Lb. paracasei subsp. paracasei. The 10 strains tolerated high concentration of bile salts by Minimal Inhibitory Concentration and growth rate assays and showed susceptibility or moderate susceptibility against antibiotics of human and veterinary importance, except for the resistanceto the class of aminoglycoside antibiotics. Eight out 10 strains showed in vitro cholesterol-lowering ability, whereas all strains showed antioxidant activity of their cell-free supernatants. Moreover, the four strains with highest survival to simulated gastrointestinal transit showed the ability to adhere to Caco-2 cells.
Conclusion: The results suggest that some strains may be effective probiotics to be use as tool to design probiotic dairy products after confirmation probiotic activities in further in vivo studies. Findings of the present study suggest that four strains showed good or strong adherent ability on Caco-2 cell monolayer, that is one of prerequisite that probiotic bacteria must have to perform their functional properties.
Keywords: adhesion, antioxidant activity, bile salts, cholesterol assimilation, dairy products, gastrointestinal resistance, probiotic lactobacilli.
Nowadays producing foods with high organoleptic and nutritional standards, stable against spoilage and safe for human health, is of a growing interest for food industries.
Over the years, as a consequence of changing lifestyles, our eating habitude have undergone significant changes, with a strengthening of the relationship between food and health. This aspect has increased consumer interest in foods that, in addition to suitable nutritional values, include microorganisms with functional properties (i.e. probiotics, antimicrobial and bioactive peptide production, antioxidant activity) able to exert positive effects on human health [1].
Probiotics are “live microorganisms which when administered in adequate amounts confer health benefits on the host” [2]. The definition does not necessarily imply that probiotic must be alive and viable. Recent literature reports that health promoting effects can also be exerted by inactivated microbial cells (paraprobiotics) or by metabolites released in cell-free supernatants (postbiotics) [3].
Main beneficial effects, some proven while others deemed only potential, include modulation of immune response, cholesterol-lowering activity, prevention/alleviation of symptoms from allergic diseases of food origin, antagonism against pathogens, treatment of inflammatory bowel disease, reduction of risk factors for colon cancer [4].
Although the beneficial effects of probiotics are and remain strain specific, the concept that similar mechanisms exist among members of certain taxonomic groups of probiotic bacteria has recently been highlighted [5]. The most common species and strains of probiotic microorganisms belong to the Lactic Acid bacteria (LAB) (mainly Lactobacillus and Enterococcus) and to the genus Bifidobacterium, although some species of Bacillus and Yeast are commonly used in probiotic preparations [4, 6]. Despite the requested human origin of probiotic strains, traditional fermented milk and cheeses represent a rich source and an excellent matrix to deliver microorganisms with potential probiotic characteristics [7]. However, potentially probiotic lactobacilli have also been isolated from fermented meat [8,9] fruit, vegetable and cereal products, also being suitable probiotic carriers [10].
The use of selected autochthonous LAB as starter and functional cultures is becoming a common practice in cheese making process. Moreover, it is known that artisanal cheeses made from raw milk without starter cultures or with natural starters (i.e. natural milk and natural whey cultures) represent the best source for the isolation of LAB strains with desired technological and functional characteristics [11].
The selection process of bacterial isolates from foods involves a series of in vitro test to assess their functional traits. Resistance to simulated gastric and pancreatic juices, as well as to bile salt and colonization of the human gastrointestinal tract are among the most important properties in a probiotic culture [4, 12]. Moreover, microbial cultures must be identified at the species and strain level and must meet specific safety requirements that identify the microorganisms as Generally Recognized As Safe (GRAS, in the US) and Qualified Presumption of Safety (QPS, in the EU) [13]. Finally, probiotic interesting microbial strains must be validated in in vivo studies, such as reproducible clinical trials on human subjects [12, 14].
The selection of autochthonous strains with appropriate functional characteristics is a topic widely explored in the current literature [15-18]. Nevertheless, the potential relevance of applicative implications makes surveys focus on this issue still worthy of continuous studies. New research can provide better knowledge for formulation of new defined functional starter cultures to be used in the production of traditional fermented functional dairy products.
Autochthonous cultures are usually selected from the food products they are going to be employed for. In a previous work, we report the dynamics of bacterial communities during manufacture and ripening of traditional Caciocavallo cheese by 16S rRNA gene pyrosequencing and viable counts [19]. These results can contribute to improving the selection process of isolated microbial cultures to be reintroduced into the same food ecosystem.
This study aimed to provide an investigation on the selection of functional relevant microbial strains of Lactobacillus spp. isolated during cheese making and ripening of Caciocavallo of Castelfranco cheese.
2.1 Bacterial isolates, culture conditions and preliminary selection of most promising strains
Lactobacillus isolates included in this study were isolated during cheese making and ripening of Caciocavallo of Castelfranco cheese as described in a previous work [19]. Briefly, aliquots of serial 10-fold dilutions of each sample were isolated on modified (m)MRS-BPB agar [De Man-Rogosa and Sharpe (MRS, Oxoid, Rodano, Milano, Italy) agar at pH 6.5, supplemented with 0.05% L-cysteine/HCl (Sigma-Aldrich, St Louis, MO, USA) and 0.002% bromophenol blue] prepared according to [20], and incubated in anaerobiosis (Anaerogen kit, Oxoid) for 48 h at 37 °C.
Colonies were purified on MRS agar and examined for cellular morphology, Gram stain, catalase (H2O2 3% v/v) and oxidase test (Oxidase Strips, Sigma-Aldrich, Milano, Italy). Pure cultures were stored at -20°C in appropriate culture media supplemented with 25% (v/v) sterile glycerol.
A total of 25 LAB, presumptively identified as Lactobacillus spp., were isolated from cheeses after 1 (C1, 5 isolates), 30 (C30, 12 isolates) and 60 (C60, 8 isolates) days of ripening. The isolates were preliminarily screened for their survival to simulated gastro-intestinal transit in order to select the most promising isolates to be assayed for others probiotic potential activities. Before each in vitro assay, the isolates were sub-cultured twice in MRS broth.
2.2 Survival to simulated gastrointestinal transit assay
Twenty-five isolates of presumptively Lactobacillus spp., were examined for their tolerance to simulated gastrointestinal transit, according to [21] with slight modifications. After growth in MRS broth for 16 hours, culture broths were centrifuged at 5000 g for 10 min and the pellets were washed in sterile Ringer’s solution (Oxoid). Two ml of cell suspensions were centrifuged at 13000 g for 5 min and resulting pellets were suspended in an equal volume of simulated saliva (SS: 86mmol L-1NaCl, 7 mmol L-1 KCl, 45 mmol L-1 NaHCO3, 100 mg L-1 lysozyme (Sigma), pH 6.90) and incubated at 37 °C for 5 min. Bacterial cells were then harvested by centrifugation and resuspended in 2 ml of simulated gastric juice (SGJ: 86 mmol L-1 NaCl, 7 mmol L-1 KCl, 45 mmol L-1 NaHCO3, 3 g L-1 pepsin, pH 2.5) and incubated at 37 °C for 120 min under constant stirring at 200 rpm in order to simulate the peristaltic movements. Bacterial suspensions were then centrifuged and the pellets resuspended in an equal volume of simulated pancreatic juice (SPJ: 22 mmol L-1 NaCl, 3.2 mmol L-1KCl, 76 mmol L-1 NaHCO3, 0.5% porcine bile (Sigma), 0.1% pancreatin (Sigma), pH 7.5) and incubated at 37 °C for 120 min at 200 rpm.
Simulated juices (SS; SGJ and SPJ), without enzymes, were sterilized at 121 °C for 15 min. Enzyme solutions were sterilized by filtering through 0.45 µm membrane syringe-filters and added to sterile simulated juices.
The isolates were counted before and at the end of simulated gastrointestinal transit. Bacterial counts were determined on MRS agar (Oxoid). Plates were incubated at 37 °C for 48 h under anaerobic conditions. Lb. rhamnosus GG (ATCC 53103) was used as reference strain.
The percentage of survival rate after the gastrointestinal transit was calculated as follows: N (log CFU/ml of isolate after incubation under the test conditions)/N0 (log CFU/ml of the assayed isolate before gastrointestinal transit) × 100.
All the isolates that showed a survival rate ≥85% were first characterized at strain and species level and then assayed for other probiotic activities.
2.3 Strain typing and species identification
DNA was extracted from overnight cultures by using InstaGeneTM Matrix (Bio-Rad, Milano, Italy) according to the manufacturer's instructions, as described previously [22].
DNA templates were used for strain typing by Repetitive Sequence-Based PCR (rep-PCR) and to identify the isolates at species level by sequencing of 16S rRNA and by species-specific PCR as described below.
The rep-PCR was performed in a total volume of 25 μl containing 50 ng of DNA template, 2.5 µl of TaqDNA polymerase 10X buffer (Invitrogen, Italy), 1.75 µl of 50 mM MgCl2, 1 µl of a deoxynucleoside triphosphate mix (10 mM each), 0.15 µl (0.1 mM) of primer (GTG)5 5’-GTGGTGGTGGTGGTG-3’ [23] and 0.5 µl of 5 U/µl Taq DNA polymerase (Invitrogen). PCR conditions consisted of an initial DNA melting (95 °C for 4 min) followed by 35 cycles (94 °C for 1 min; 40 °C for 1 min; 72 °C for 1 min) and a final extension cycle at 72 °C for 8 min. PCR amplicons were run in 1.75% agarose gel stained with 0.01% SYBR Safe DNA gel stain (Invitrogen) at 100 V for 2 h. DNA-fingerprintings profiles were visualized with UV trans-illuminator UVIdocHD2 (UVITEC, Cambridge, UK) and were analysed by the Bionumerics software (version 5.1, Applied Maths).
Amplification of 16S rRNA gene was performed by using primers fD1 (5’-AGAGTTTGATCCTGGCTCAG-3’) and rD1 (5’-AAGGAGGTGATCCAGCC-3’) (Escherichia coli positions 8-17 and 1540-1524, respectively) [24]. PCR conditions were previously described [19], and PCR products were run in agarose gel (1.5% w/v) for 90 min at 100V and then purified by using QIAquick PCR purification kit (Qiagen S.p.A., Milan) following the manufacturer’s protocol. Sequencing data of 16S were viewed using PC FinchTV software (http://www.geospiza.com/finchtv/) and DNA similarity analysis was performed using the BLAST (bl2seq) program at the NCBI website (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Multiplex PCR was performed by species-specific primers designed by [25] with minor modifications. Amplifications were performed in a total volume of 50 μl containing 20 mMTris-HCl, 50 mMKCl, 1.5 mM MgCl2, deoxynucleoside triphosphate mix (500 µM each), 10 pmol each of primers PAR (5’-GACGGTTAAGATTGGTGAC-3’), CAS (5’-ACTGAAGGCGACAAGGA-3’), and RHA (5’-GCGTCAGGTTGGTGTTG-3’), 50 pmol of primer CPR (5’-CAANTGGATNGAACCTGGCTTT-3’), 2.5 U of TaqDNA polymerase (Invitrogen, Italy), and 100 ng of template DNA. PCR conditions consisted of an initial template melting (95 °C for 5 min) followed by 30 cycles (95 °C for 30 s; 52 °C for 1 min; 72 °C for 1.5 min) and one elongation cycle for 7 min at 72 °C. PCR amplicons were separated by agarose (2%, w/v) gel electrophoresis at 100 V for 1 h and then visualized with UV trans-illuminator UVI doc HD2 (Uvitech, UK).Lb. rhamnosus GG (ATCC 53103), Lb. casei Shirota and Lb. paracasei LMG P-21380, were used as reference strains.
2.3.1. Bile salts tolerance and Bile Salt Hydrolase (BSH) activity
Ten strains of Lb. paracasei subsp. paracasei selected on the basis of their ability to survive to simulated gastrointestinal juices were assayed for bile salts tolerance and bile salt hydrolase activity (BSH).
The Minimal Inhibitory Concentration (MIC) of a mix of bile salts (Oxgall, Sigma-Aldrich) and of taurocholic acid sodium salt (TCA), sodium taurodeoxycholate (TDCA), glycocholic acid sodium salt (GCA) and sodium glycodeoxycholate (GDCA) (Sigma-Aldrich) was evaluated as described previously [8]. Briefly, 10 µl of overnight culture (about 107 CFU/ml) of each strain were spotted on MRS agar plates containing bile salts mix and a single salt (0%, 0.1%, 0.2%, 0.3%, 0.4% and 0.5%). Plates were incubated anaerobically at 37 °C for 4 days. The MIC was defined as the lowest concentration of bile salts that causes a total inhibition on the growth of the colonies in the spots. The test was carried out in duplicate for all the isolates and for each type of bile salts.
The growth kinetics of the selected strains was also monitored in MRS broth containing 0.2% sodium thioglycollate and supplemented with 0.0 (MRS-THIOcontrol) or 0.2 or 0.3% of oxgall (MRS-THIOox), according to [26]. Bacterial growth was monitored at 37 °C every hour for 48 h and expressed as the time (hours) required to increase the absorbance (590 nm) of 0.3 units in MRS-THIO broth without oxgall (0%) or with 0.2% or 0.3% of oxgall. The difference in hours between the control and the test culture to increase the absorbance of 0.3 units was considered as the lag time (LT). For both MIC and growth rate assays, Lb. rhamnosus GG was used as reference strain.
Bile Salt Hydrolase (BSH) activity was assayed according to [27] with minor modifications. MRS agar plates with and without 0.3% of TDCA (pre-reduced anaerobically at 37 °C for 24 h), were streaked with overnight culture of the strains and anaerobically incubated at 37 °C for 48-72 h. The presence of a white precipitate around colonies indicated BSH activity.
2.4 Cholesterol removal
Cholesterol assimilation by 10 strains of Lb. paracasei subsp. paracasei was assessed by adding 300 µg/ml of water-soluble cholesterol PEG-600 (Sigma-Aldrich) in the following media: MRS broth and simulated intestinal fluid (0.85% (w/v) NaCl, 6.8 g/L potassium phosphate monobasic, 1.5 g/L Oxgall, 3.5 g/L glucose, and 10 g/L pancreatin, pH 6.8) according to [28]. A stock solution (3 mg/ml) of water-soluble cholesterol PEG-600 (Sigma-Aldrich) was prepared, sterilized through 0.22 µm filter (Millipore, Corp., Bedford, MA, USA) and added to the media to obtain a final concentration of 300 µg/ml of cholesterol. Overnight cultures of strains of Lb. paracasei subsp. paracasei and of Lb. rhamnosus GG (as reference strain) were inoculated at 1% (v/v) in each cholesterol-medium and incubated at 37 °C for 24 hours under anaerobic conditions. Cholesterol concentrations before and after incubation were determined according to o-phthalaldehyde (OPA, Sigma) method [28, 29] by reading the absorbance of the mixtures at 550 nm using UV-spectrophotometer (BioSpectrometer, Eppendorf). A standard curve of absorbance was prepared using cholesterol concentrations between 0 to 500 µg/ml (R2=0.9979). The ability of the strains to assimilate cholesterol was determined as follows:
%cholesterol assimilated = [cholesterol (µg/ml)0h cholesterol (µg/ml)24h/cholesterol (µg/ml)0h] × 100
2.5 Antibiotic susceptibility
Antibiotic susceptibility of the 10 strains of Lb. paracasei subsp. paracasei was assessed by the disk diffusion method [30, 31]. Petri dishes (90 mm diameter), containing 15 ml of solidified MRS agar, were overlaid with 5 ml of MRS agar soft (0.75% agar, w/v) seeded with 0.2 ml of overnight MRS broth cultures standardized to an optical density at 590 nm of 0.1. After solidification, disks were placed on the plates with the following antibiotics (Oxoid, Italy): gentamycin (30 μg), chloramphenicol (10 μg), tetracycline (30 μg), erythromycin (5 μg), clindamycin (2 μg), streptomycin (25 μg), kanamycin (30 μg) and ampicillin (10 μg). The plates were incubated anaerobically at 37 °C for 24 h. After incubation, the diameter (mm) of inhibition zone was measured. The strains were classified as resistant (R), moderately sensitive (MS), and sensitive (S) on the basis of the diameters of inhibition zones. Lb. rhamnosus GG was used as a reference strain.
2.6 Antioxidant activity
The antioxidant activity of the 4 strains of Lb. paracasei subsp. paracasei was determined on both intact cell suspensions and on the supernatant of coagulated skim milk cultures.
For the preparation of cell suspension (CS), overnight cultures of each strain where centrifuged at 5000 x g for 10 min at 4 °C. Cell pellets were washed twice in phosphate-buffered saline (PBS: 0.80% NaCl, 0.02% KCl, 0.02% KH2PO4, and 0.22% Na2HPO4, pH 7.4) and finally suspended in the same buffer at a concentration of about 109 cells/ml.
Cell-free supernatants (CFS) were prepared according to [32] with some modifications. Overnight cultures of each strain where inoculated at 2% in 10% (w/v) of sterilized reconstituted skim milk (RSM, Oxoid) supplemented with 1% glucose and 0.5% yeast extract and incubated for 24 h at 37 °C. Subsequently each milk culture was inoculated at 2% in RSM, incubated for 24 h at 37 °C and centrifuged at 5000 xg for 10 min a 4 °C. The resulting supernatant was adjusted to pH 7.4 and centrifuged again at 13000 x g for 10 min at 4 °C, after which it was sterilized with a 0.22 µm filter membrane.
To measure the antioxidant ability of the Lactobacillus strains, the DPPH (α,α-diphenyl-β-picrylhydrazyl) free radical scavenging method was used [26]. A 445 µl aliquot of CS or CFS and 555 µl of DPPH solution (0.2 mM in ethanol) were mixed in a cuvette and allowed to react for 30 min in the dark. PBS or supernatant at pH 7.4, prepared by acidifying RSM with lactic acid, were used as control samples replacing CS and CFS, respectively. The scavenged DPPH was monitored by measuring the decrease in absorbance at 517 nm. The radical scavenging activity was calculated as percentage inhibition by using the following equation:
% inhibition = (Ac – As)/Ac x 100
where Ac is the absorbance in the control sample and As is the absorbance in the sample.
2.7 Adhesion of bacterial strains to Caco-2 Cells
The human intestinal Caco-2/TC7 cell line was provided by Monique Rousset (Institute National de la Santé et de la Recherche Médicale, INSERM, Paris, France) and routinely maintained as described by [33]. For the adhesion assay, Caco-2 cells were seeded in 12-well plates (Becton Dickinson, Milan, Italy) and, after confluency, were left for 14-17 days to allow differentiation [34]. Medium was changed three times a week. Complete DMEM was replaced with antibiotic- and serum- free DMEM 16 h before the assay. Four strains with highest survival to simulated gastrointestinal transit (FI3L1T, FF3L17T, FF3L16M and FFL21T) were choose to test adhesion ability. On the day of the assay, overnight bacterial cultures of each Lb. paracasei subsp. paracasei strains and Lb. rhamnosus GG were diluted 1:100 in MRS broth and grown to the exponential growth phase. After monitoring the OD600, appropriate amounts of bacterial cells were harvested by centrifugation at 5000× g for 10 min, resuspended in antibiotic- and serum-free DMEM and added to cell monolayers at a concentration of 1×108 CFU/well. Co-cultures of bacteria and Caco-2 cells were incubated at 37°C for 1.5 h. Non-adhering bacteria were then removed by 5 washes with Hanks’ Balanced Salt solution (HBSS: 137 mmol L−1NaCl, 5.36 mmol L−1KCl, 1.67 mmol L−1 CaCl2, 1 mmol L−1 MgCl2, 1.03 mmol L−1 MgSO4, 0.44 mmol L−1KH2PO4, 0.34 mmol L−1 Na2HPO4, 5.6 mmol L−1 glucose) and cell monolayers were lysed with 1% Triton-X-100, according to [35]. Adhering, viable bacterial cells were quantified by plating appropriate serial dilutions of Caco-2 lysates on MRS agar medium.
2.8 Statistical analysis
In vitro adhesion experiments were performed at least in triplicate. Data are presented as mean ± SD. Prior to analysis, normal distribution and homogeneity of variance of all variables were assumed with Shapiro–Wilk and Levene’s tests, respectively. Statistical significance was evaluated by Kruskal-Wallis test for equal medians. Statistical analysis was performed with “Past” software package (version 3.13). Differences with p values < 0.05 were considered significant.
In vitro probiotic and functional tests were performed in duplicate in three independent experiments.
Statistical analysis was performed using Stata software package version 13 (Stata, 2013). All data are expressed as means ± standard deviation (SD) and were analysed by one-way analysis of variance (ANOVA). Comparisons of mean results were carried by Tukey’s post-hoc test at p ≤0.05 significance level.
3.1 Survival under simulated gastrointestinal tract conditions, molecular identification and characterization of strains and bile salts tolerance
Promising probiotic bacteria should be able to survive in the hostile gastrointestinal environment. Gastric and intestinal juices act as the highest hurdles for the survival of ingested microorganisms and resistance to low pH and bile salt is considered a prerequisite for selecting potential probiotics [36].
In the present work, only the isolates that showed a survival rate to simulated gastrointestinal transit ≥85%were selected for further study. As already reported in the current literature [21], these values could be a proper range to select the bacteria resistant to gastrointestinal conditions.
In this study, after the consecutive passage in salivary juice (5 min, pH 6.9), in gastric juice (120 min, pH 2.5) and in duodenal juice (120 min, pH 7.5), 16 out of 25 isolates of Lactobacillus spp. showed a survival rate ≥85%. The results are shown in Table 1. The highest survival rate is presented by the strains FFL21T and FF3L16M (% survival of 98.90 and 97.76, respectively) and comparable to the reference probiotic strain Lb. rhamnosus GG, which was the most resistant strain. rep-PCR analysis of the 16 isolates resulted in 10 different profiles (Table 1; Figure 1), whereas 16S rRNA gene sequencing revealed high similarity (99%; accession number: NC014334.2/NCPZ013921.1) to closely related Lb. casei and Lb. paracasei subsp. paracasei species, as widely reported in the literature [37, 38]. However, in this study, tuf multiplex PCR assay (Table 1) allow species-specific identification of the 10 strains as Lb. paracasei subsp. paracasei species by generating two distinct amplicons of 240 bp and 520 bp, as the reference strain Lb. paracasei subsp. paracasei LMG P-21380 (Figure 2, line P). The strain Lb. casei Shirota showed three bands (700, 520 and 350 bp) as expected (Figure 2, line C), whereas Lb. rhamnosus GG generated one PCR product of about 520 bp (Figure 2, line GG). All strains of Lb. paracasei subsp. paracasei were isolated during cheese ripening (1, 30 and 60 days) of traditional Caciocavallo of Castelfranco cheese where the Lb. casei group becomes the subdominant population at the end of ripening as detected by 16S rRNA gene pyrosequencing [19]. According to our results, [39] found that among several Non-Starter LAB (NSLAB) strains isolated from two type of cheese and identified by 16S rRNA gene sequencing, the most resistant strains to gastrointestinal transit belonged to Lb. casei/Lb. paracasei group. Moreover, Lb. paracasei subsp. paracasei was found in the human intestine and recognized as a probiotic [40].
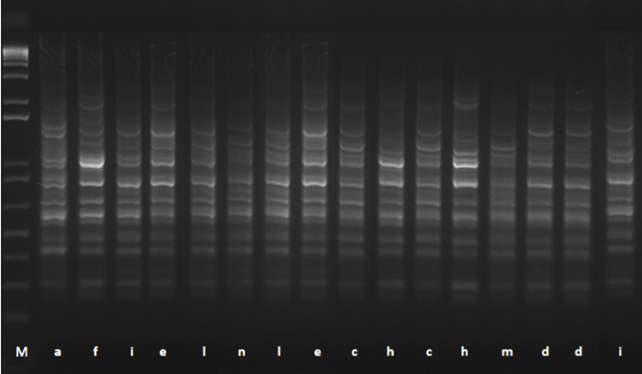
Figure 1. DNA-fingerprintings of 16 isolates of lactobacilli with a survival rate ≥85% after exposure to simulated gastrointestinal juices. Different strains are indicated with different letters (from a to n) and correspond to the strains reported in the Table 1. M: molecular marker 1 Kb plus DNA Ladder (Invitrogen, Italy).
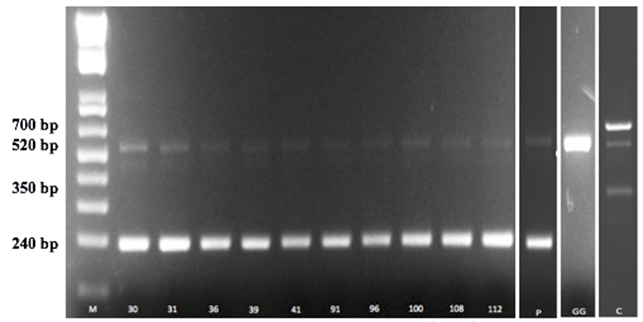
Figure 2. Amplification products obtained from the tuf multiplex assay. Lane M, 1 Kb plus DNA ladder (Invitrogen, Italy). Numeric codes of each strain profile corresponding to the strains reported in the Table 1. lane P, Lb. paracasei subsp. paracasei LMG P-21380; Lane GG, Lb. rhamnosus GG; Lane C, Lb. casei Shirota.
Table 1. Strain differentiation, molecular identification and survival rates of strains of Lactobacillus paracasei after exposure to simulated gastric juices
|
Strains |
Samplea |
rep-PCR-profileb |
tuf multiplex assay |
N0 (Log CFU/ml)e |
N (Log CFU/ml)e |
% survivale |
||
|
Speciesc |
Profiled |
|||||||
|
FF3L13T |
C30 |
a |
Lactobacillus paracasei |
30 |
9.11±0.18 |
7.82±0.13 |
85.83 |
|
|
FF6L7T |
C60 |
f |
Lb. paracasei |
31 |
8.89±0.08 |
7.68±0.03 |
86.38 |
|
|
FF3L14T |
C30 |
c |
Lb. paracasei |
ND |
8.55±0.18 |
7.53±0.02 |
88.07 |
|
|
FF3L4T |
C30 |
c |
Lb. paracasei |
91 |
8.77±0.09 |
7.74±0.04 |
88.25 |
|
|
FF6L3T |
C30 |
l |
Lb. paracasei |
ND |
8.73±0.19 |
7.81±0.12 |
89.46 |
|
|
FF3L3M |
C30 |
l |
Lb. paracasei |
39 |
8.93±0.02 |
8.02±0.08 |
89.80 |
|
|
FI3L12T |
C30 |
e |
Lb. paracasei |
ND |
8.74±0.23 |
8.08±0.13 |
92.44 |
|
|
FF6L8T |
C60 |
m |
Lb. paracasei |
100 |
8.73±0.03 |
8.21±0.04 |
94.04 |
|
|
FF6L17T |
C60 |
d |
Lb. paracasei |
ND |
8.83±0.05 |
8.31±0.11 |
94.11 |
|
|
FF3L6M |
C30 |
n |
Lb. paracasei |
41 |
9.04±0.12 |
8.52±0.18 |
94.24 |
|
|
FF3L13M |
C30 |
i |
Lb. paracasei |
ND |
8.61±0.22 |
8.19±0.14 |
95.12 |
|
|
FF3L17T |
C30 |
d |
Lb. paracasei |
108 |
8.74±0.11 |
8.37±0.08 |
95.76 |
|
|
FI3L1T |
C30 |
i |
Lb. paracasei |
112 |
8.63±0.05 |
8.33±0.06 |
96.52 |
|
|
FF3L16M |
C30 |
e |
Lb. paracasei |
36 |
8.51±0.10 |
8.32±0.01 |
97.76 |
|
|
FIL2M |
C1 |
h |
Lb. paracasei |
ND |
8.91±0.12 |
8.78±0.19 |
98.54 |
|
|
FFL21T |
C1 |
h |
Lb. paracasei |
96 |
8.24±0.02 |
8.15±0.09 |
98.90 |
|
|
GGf |
- |
- |
Lb. ramnhosus |
GG |
9.10±0.12 |
9.11±0.08 |
100.1 |
|
|
Shirotaf |
- |
- |
Lb. casei |
C |
- |
- |
- |
|
|
LMG P-21380f |
- |
- |
Lb. paracasei |
P |
- |
- |
- |
|
a:C1, cheese after one day of ripening; C30, cheese after 30 days of ripening; C60, cheese after 60 days of ripening; b: rep-PCR profile reported in Figure 1; c: species identification after Multiplex PCR; all the strains were identified by 16SrRNA sequencing as Lb. casei/paracasei; d: profile of identified specie is reported in Figure 2; ND: not determined; e:survival % = [N/N0]×100 where N represents the log CFU/ml of strains after incubation under the test conditions and N0 represents the log CFU/ml of the assayed strain before gastrointestinal transit; results are mean of six replicates ± standard deviation; f: Lb. rhamnosus GG, Lb. casei Shirota and Lb. paracasei LMG P-21380 were used as reference strains.
The effects of bile salts on the survival of the 10 strains of Lb. paracasei subsp. paracasei was performed by measuring the MIC against increasing concentrations (from 0.0 to 0.5%) of a mixture of bile salts (Oxgall) or of single bile acids (TDCA, TCA, GCA, GDCA). Literature data report that in the human gastrointestinal tract the mean bile concentration is about 0.3% (w/v) and is considered as critical and high enough to screen for resistant strains [8, 41]. The results of bile salts tolerance are shown in Table2.
Table 2. Bile salts minimal inhibitory concentration (MIC) of strains of Lactobacillus paracasei
|
Species |
Strain |
MICa |
|
Growth in MRS-thioglycollate broth with oxgall (%) |
||||||||||||||||||||||
|
Oxgall |
TCA |
TDCA |
GCA |
GDCA |
|
0% |
|
0.2% |
|
0.3% |
||||||||||||||||
|
|
h0.3b |
|
h0.3b |
LTc |
|
h0.3b |
LTc |
|||||||||||||||||||
|
Lb. paracasei |
FF3L13T |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
|
7 |
|
11 |
4a |
|
13 |
6e |
||||||||||||
|
Lb. paracasei |
FF6L7T |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
5 |
|
7 |
2b |
|
7 |
2b |
|||||||||||||
|
Lb. paracasei |
FF3L4T |
>0.5 |
>0.5 |
> 0.5 |
> 0.5 |
>0.5 |
5 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FF3L3M |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
10 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FF6L8T |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
8 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FF3L6M |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
8 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FF3L17T |
> 0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.1 |
9 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FI3L1T |
0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
9 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FF3L16M |
> 0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
9 |
|
> 48 |
> 48d |
|
> 48 |
> 48d |
|||||||||||||
|
Lb. paracasei |
FFL21T |
0.4 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
5 |
|
5 |
0c |
|
5 |
0c |
|||||||||||||
|
Lb. rhamnosus |
GG |
> 0.5 |
> 0.5 |
> 0.5 |
> 0.5 |
0.2 |
4 |
|
5 |
1b |
|
7 |
3ab |
|||||||||||||
a: MIC, as the lowest concentration (%) of bile salts that causes a total inhibition on the growth of the colonies in the spots. TCA, taurocholic acid sodium salt; TDCA, sodium taurodeoxycholate; GCA, glycocholic acid sodium salt; GDCA, sodium glycodeoxycholate (Sigma-Aldrich). b: time (h) required to increase the absorbance (600 nm) of 0.3 units in MRS-THIO broth without oxgall (0%) or with 0.2% or 0.3% of oxgall. c: LT, latency time that is the delay (h) of the bacterial growth in presence of oxgall calculated as the difference between the time (h) required to increase the absorbance (600 nm) of 0.3 units with oxgall (0.2% or 0.3%) and the time required to increase the absorbance (600 nm) of 0.3 units without oxgall (0%). Values within the columns with different superscript letters (a-e) differ at P <0.05.
The 10 strains of Lb. paracasei subsp. paracasei were able to tolerate concentrations greater than 0.5% of TCA, TDCA and GCA, whereas, with the exception of strain FF3L4T, the MIC of GDCA was of 0.1-0.2% for all the strains, suggesting that GDCA was the limiting bile acid at which the strains were more sensible. The tolerance to oxgall was variable among the strains, with MICs ranging from 0.4 to >0.5%. Moreover, growth kinetics, with and without 0.2 and 0.3% of oxgall, were determined for the 10 strains. As shown in the Table 2, 7 out of 10 strains exhibited latency time above 48 h. Only the strain FFL21T did not show delay on growth when incubated with oxgall compared to the control, while the reference strain Lb. rhamnosus GG showed a latency times of 1 and 3 h at concentration of 0.2% and 0.3% of oxgall, respectively. According to our results, several authors highlight that bile salt tolerance may be strain-specific [26, 42]. The 10 strains of Lb. paracasei subsp. paracasei were also assayed for the presence of Bile Salt Hydrolase (BSH) enzyme. All the strains did not show precipitation zones around the colonies or growth of opaque colonies on MRS agar with TDCA, suggesting the lack ability to deconjugate bile salts (data not shown). Some LAB strains, although not carrying BSH activity, exhibited high tolerance to bile salts. Studies by [43] showed that the resistance to bile salts by lactobacilli may not be related to BSH activity as also showed by [44] that found NSLAB strains able to tolerate bile salts without the ability to produce BSH.
3.2 Cholesterol-lowering activity
An excess of serum cholesterol is associated with cardiovascular disease in human [45]. Many in vitro and/or in vivo studies have demonstrated the cholesterol-lowering activity of probiotic LAB strains [27, 46-50], even if the mechanisms of this reduction are still partially unknown [45].
The results of the in vitro cholesterol-lowering ability of the 10 strains of Lb. paracasei subsp. paracasei in MRS broth containing 300 μg/ml of cholesterol are reported in Table 3. The percentage of cholesterol removed by the strains ranged from 10.40% (strain FF3L17T) to 26.29% (strain FF3L16M). Two strains (FI3L1T and FF3L6M) were not able to remove cholesterol in the medium, whereas, the reference strain Lb. rhamnosus GG removed 35.30% of the cholesterol initially added in the medium. These results are in agreement with [28] who report removal rates between 13 and 39% by 11 lactobacilli strains grown in MRS broth with 100 ?g/ml of cholesterol. Moreover, the same strain of Lb. rhamnosus GG tested in our study showed 29.98% of removal rates.
Table 3. Cholesterol removal ability by strains of Lactobacillus (Lb.) paracasei in MRS broth, MRS broth supplemented with 0.3% of oxgall and in simulated intestinal fluid.
|
Species |
Strains |
Cholesterol assimilated (%)a ± SD |
|
|
MRS broth |
Simulated intestinal fluid |
||
|
Lb. paracasei |
FI3L1T |
0.00±0.70 |
NDb |
|
Lb. paracasei |
FF3L6M |
0.00±1.30 |
ND |
|
Lb. paracasei |
FF3L17T |
10.40±0.94 |
ND |
|
Lb. paracasei |
FF6L8T |
15.60±5.30 |
ND |
|
Lb. paracasei |
FF3L4T |
16.10±7.60 |
ND |
|
Lb. paracasei |
FF6L7T |
17.79±3.58 |
51.20±0.03 |
|
Lb. paracasei |
FF3L3M |
20.00±2.30 |
ND |
|
Lb. paracasei |
FF3L13T |
21.90±2.20 |
9.39±9.40 |
|
Lb. paracasei |
FFL21T |
25.30±6.00 |
15.85±3.10 |
|
Lb. paracasei |
FF3L16M |
26.29±6.30 |
ND |
|
Lb. rhamnosus |
GG |
35.30±16.7 |
56.32±4.33 |
a: % cholesterol assimilation = [cholesterol (μg/ml)0h- cholesterol (μg/ml)24h/cholesterol (μg/ml)0h] × 100. The results are the mean of % cholesterol assimilated by the strains ± sd; b: Not Determined.
Cholesterol-lowering ability was also determined in simulated intestinal fluid added of 300 μg/ml of cholesterol. The assay was performed only for the 3 strains that showed a fast growth in MRS-THIO broth with 0.3% of oxgall (see Table 2). The results reported in Table 3 indicated that1 out of 3 strains of Lb. paracasei subsp. paracasei and the strain of Lb. rhamnosus GG, presented a higher rate of removal in simulated intestinal fluid than in MRS broth. These results suggest that the cholesterol assimilation may be influenced by the growth media composition and, in particular, as already reported in the literature, the addition of bile salts increases the amount of cholesterol assimilated [28, 45, 51].
3.3 Antibiotic susceptibility
The results of the susceptibility of the 10 strains of Lb. paracasei against 8 antibiotics of human and veterinary importance [52] are reported in Table 4. All the strains were sensitive to ampicillin (AMP), tetraclycine (TE), gentamycin (CN), and moderately sensitive to clindamycin (DA) and, except for sensitive strain FF3L4T to chloramphenicol (C). Moreover, all strains, including the reference strain Lb. rhamnosus GG, showed resistance to the aminoglycoside inhibitor of protein synthesis kanamycin (K), whereas the strains FF3L9T and FI3L1T were resistant to streptomycin (S) and the strain FF3L4T to erythromycin (E).
Our results are generally in agreement with the data reported previously on the antibiotic susceptibility of strains of Lb. paracasei isolated from traditional Serbian cheese[53] and Parmigiano Reggiano cheese [44].
Safety concerns on the use of antibiotic resistant strains as probiotic arise from the possibility that antibiotic resistant genes could be transmitted to intestinal pathogenic bacteria [54]. The determination of antibiotic resistant genes location could contribute in safely application of antibiotic resistant probiotic LAB [53]. Particularly, the resistance of our strains to the class of aminoglycoside antibiotics (such as streptomycin and kanamycin) is widespread among lactobacilli [55] and considered to be non-transmissible [56].
Table 4. Antibiotic susceptibility of strains of Lactobacillus (Lb.) paracasei determined by agar disk diffusion method
|
Species |
Strains |
Antibioticsa |
|||||||||||||||||||||||
|
AMP |
|
K |
|
DA |
|
TE |
|
CN |
|
C |
|
S |
|
E |
|||||||||||
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|
Db |
Sc |
|||
|
Lb. paracasei |
FF3L13T |
20 |
S |
0 |
R |
12 |
MS |
17 |
S |
7 |
S |
10 |
MS |
1 |
MS |
19 |
MS |
||||||||
|
Lb. paracasei |
FF6L7T |
18 |
S |
0 |
R |
22 |
MS |
16 |
S |
6 |
S |
10 |
MS |
1 |
MS |
17 |
MS |
||||||||
|
Lb. paracasei |
FF3L4T |
26 |
S |
0 |
R |
20 |
MS |
21 |
S |
14 |
S |
20 |
S |
8 |
S |
3 |
R |
||||||||
|
Lb. paracasei |
FF3L3M |
28 |
S |
0 |
R |
15 |
MS |
20 |
S |
5 |
S |
10 |
MS |
1 |
MS |
20 |
MS |
||||||||
|
Lb. paracasei |
FF6L8T |
23 |
S |
0 |
R |
18 |
MS |
18 |
S |
6 |
S |
10 |
MS |
1 |
MS |
19 |
MS |
||||||||
|
Lb. paracasei |
FF3L6M |
23 |
S |
0 |
R |
14 |
MS |
17 |
S |
5 |
S |
12 |
MS |
1 |
MS |
14 |
MS |
||||||||
|
Lb. paracasei |
FF3L17T |
27 |
S |
0 |
R |
24 |
MS |
22 |
S |
7 |
S |
15 |
MS |
1 |
MS |
17 |
MS |
||||||||
|
Lb. paracasei |
FI3L1T |
30 |
S |
0 |
R |
16 |
MS |
19 |
S |
5 |
S |
13 |
MS |
0 |
R |
17 |
MS |
||||||||
|
Lb. paracasei |
FF3L16M |
26 |
S |
0 |
R |
17 |
MS |
16 |
S |
6 |
S |
10 |
MS |
1 |
MS |
15 |
MS |
||||||||
|
Lb. paracasei |
FFL21T |
24 |
S |
0 |
R |
17 |
MS |
19 |
S |
7 |
S |
14 |
MS |
2 |
MS |
21 |
MS |
||||||||
|
Lb. rhamnosus |
GG |
11 |
S |
0 |
R |
12 |
MS |
15 |
S |
7 |
S |
10 |
MS |
5 |
MS |
14 |
MS |
||||||||
a: AMP, ampicillin (10 µg); K, kanamycin (30 µg); DA, clindamycin (2 µg); TE, tetraclycine (30 µg); CN, gentamycin (30 µg); C, chloramphenicol (10 µg); S, streptomycin (25 µg); E, erythromycin (5 µg); b: diameters of zones of inhibition in mm; c: Susceptibility: R (resistant), MS (moderately susceptible), or S (susceptible); c: Lb. rhamnosus GG used as a reference strain.
3.4 Antioxidant activity by DPPH radical inhibition
Antioxidant activity of L. paracasei strains was evaluated by DPPH radical scavenging method on cell free supernatants (CFS) and on cell suspension (CS) after growing the strains in RSM and MRS broth, respectively. Table 5 shows that all the CS samples provided DPPH inhibition values between 4.9% and 18.1% with the strain FF3L6M showing the highest value. The antioxidant activity of CFS samples ranged between 25.90% and 57.30% and it was significantly higher than CS samples. Nevertheless, seven CFS of Lb. paracasei, including Lb. rhamnosus GG, showed no antioxidant activity. Moreover, the antioxidant activity was significantly (P value < 0.05) higher compared to CS samples. The strain FF3L13T had the highest DPPH scavenging ability (57.3%), followed by FI3L1T (45%), FF3L6M (42.65%), and FF6L7T (25.90%). Most of the literature data report that the antioxidant activity of LAB strains, including Lb. rhamnosus GG, was higher for intact cells than that shown by cell-free extracts [26, 57, 58].
Table 5. Scavenging DPPH rate of Cell-Free Supernatants (CFS) and Cell Suspension (CS) of strains of Lactobacillus (Lb.) paracasei
|
Species |
Strains |
a% Inibition (CFS) |
a% Inibition (CS) |
|
Lb. paracasei |
FF3L13T |
57.30±1.70 |
11.35±0.21 |
|
Lb. paracasei |
FF6L7T |
25.90± 2.40 |
9.75±1.34 |
|
Lb. paracasei |
FF3L4T |
0 |
9.55±0.91 |
|
Lb. paracasei |
FF3L3M |
0 |
13.45±2.82 |
|
Lb. paracasei |
FF6L8T |
0 |
10.00±1.83 |
|
Lb. paracasei |
FF3L6M |
42.65±7.70 |
18.10±0.42 |
|
Lb. paracasei |
FF3L17T |
0 |
4.90±4.25 |
|
Lb. paracasei |
FI3L1T |
45.00± 4.52 |
7.35±5.59 |
|
Lb. paracasei |
FF3L16M |
0d |
13.90±2.12 |
|
Lb. paracasei |
FFL21T |
0 |
9.15±2.90 |
|
Lb. rhamnosus |
GG |
0 |
3.45± 2.75 |
a: % scavenging DPPH inhibition. The results are reported as mean ± sd of two replicates.
3.5 Adhesion of bacterial strains to Caco-2 Cells
The four strains with highest survival to simulated gastrointestinal transit were analyzed for their ability to adhere to Caco-2 cells, representing a suitable in vitro model of human intestinal epithelium. For each strain, bacterial cells were co-cultured with intestinal cells at an initial amount of 1×108 CFU/ml and the resulting adhering bacteria were counted on MRS agar medium. The results showed that all the tested strains were able to adhere to Caco-2 cells with similar efficiencies to that observed for Lb. rhamnosus GG probiotic control (Figure 3). In fact, about 1 × 106 – 1 x 107 CFU/ml were recovered. To better compare the results, the values were expressed as [(logCFUtf-logCFUt0)/logCFUt0]x100, representing the percentage reduction of adhering cells, in relation to their initial titer, for each strain (Figure 4). Statistical analysis revealed no significant differences among the strains, including Lb. rhamnosus GG control. Taken together, the results suggest good adhesion capacity displayed by the bacterial strains.
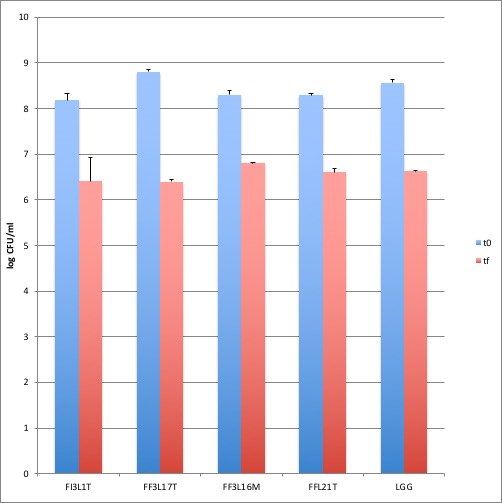
Figure 3. Adhesion to Caco-2 cells. Cell counts of viable bacterial strains and LGG adhering on differentiated Caco-2 cells co-cultured with 1 × 108 bacterial colony forming units (CFU)/well. Blu columns refer to the initial bacterial load (to), while red columns refer to adhering bacteria recovered at the end of co-incubation (tf). Data are reported as log of bacterial CFU recovered after plating. Columns represent the mean ± SD of one experiment performed in triplicate.
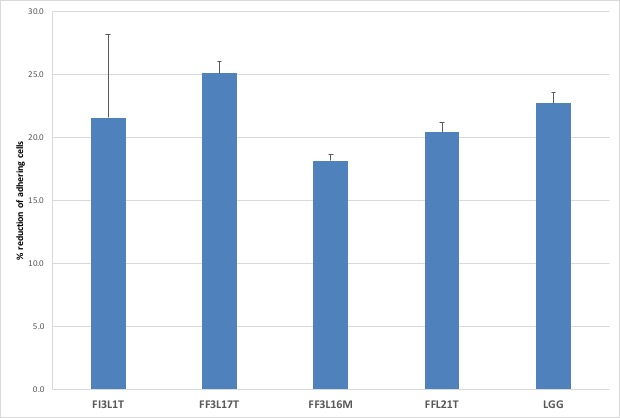
Figure 4: Reduction of bacterial strains to Caco-2 Cells. Cell counts of viable bacterial strains on differentiated Caco-2 cells. Columns represent the mean ± SD of one experiment performed in triplicate. Data are reported as [(logCFUtf-logCFUt0)/logCFUt0]x100.
Lb. paracasei subsp. paracasei and related species are among the dominant group of NSLAB during cheese ripening. Moreover, many strains have unique properties that can provide benefits for human and animal health.
In this study,10selected strains of Lb. paracasei subsp. paracasei isolated from Caciocavallo cheese were subjected to a multi-step of in vitro analyses to assess their probiotic potential properties.
The 10 strains showed a survival rate to simulated gastrointestinal transit ≥85% with the strains FFL21T and FF3L16M showing the highest values. Four out of 10 strains (FF3L17T, FI3L1T, FF3L16M, FFL21T) showed adhesion capability to Caco-2 cells from 75% to 80% and lack of undesirable traits such as non-transmissible antibiotic resistance and 2 of these strains (FF3L16M, FFL21T) had cholesterol assimilation values of about 25%. Moreover, 2 strains (FF3L13T, FF6L7T) had a good ability to assimilate cholesterol and, together with the strain FF3L6M, a high antioxidant activity of cell free supernatants after growing in reconstituted skimmed milk which could act as postbiotics.
In conclusion, our results provide evidence that Caciocavallo cheese is a good source of LAB with probiotic potential. The in vitro properties of some strains encourage further in vivo studies to improving information on their use as functional cultures in dairy products.
This study was supported by Department of Agricultural Sciences, University of Naples Federico II.
Author's contributions
M. and A.L.S. performed the experiments. P.Z. and B. G. performed Caco-2 Cells experiments F.V. was responsible for the research design and interpretation of data. All authors contributed in drafting and revising the manuscript.
The authors declare no competing interests.
Macori G, Cotter PD (2018) Novel insights into the microbiology of fermented dairy foods. Curr Opin Biotechnol 49:172-178. PMid:28964915
View Article PubMed/NCBIHill C, Guarner F, Reid G, Gibson GR, Merenstein DJ, Pot B, Morelli L, Berni Canani R, Flint HJ, Salminen S, Calder PC, Sanders ME (2014) Expert consensus document: the international scientific association for probiotics and prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat Rev Gastroenterol Hepatol 11:506-514. doi:10.1038/nrgastro.2014.66. PMid:24912386
View Article PubMed/NCBIAguilar-Toalá JE, Garcia-Varela R, Garcia HS, Mata-Haro V, González-Córdova AF, Vallejo-Cordoba B, Hernández-Mendoza A (2018) Postbiotics: An evolving term within the functional foods field. Trends Food Sci Technol 75:105-114.
View ArticleFleet M, Rahman PKSM (2018) Probiotics and Their Health Benefits (pp. 267-279). Microbial Functional Foods and Nutraceuticals, First Edition. Edited by Gupta VK, Treichel H, Shapaval V, de Oliveira LA, Tuohy MG. John Wiley & Sons Ltd.
View ArticleSanders ME, Benson A, Lebeer S, Merenstein DJ, Klaenhammer TR (2018) Shared mechanisms among probiotic taxa: implications for general probiotic claims. Curr OpinBiotechnol 49:207-216. PMid:29128720
View Article PubMed/NCBIPennacchia C, Blaiotta G, Pepe O, Villani F (2008) Isolation of Saccharomyces cerevisiae strains from different food matrices and their preliminary selection for a potential use as probiotics. J Appl Microbiol 105:1919-1928. ISSN 1364-5072. PMid:19120638
View Article PubMed/NCBIRanadheera CS, Vidanarachchi JK, Rocha RS, Cruz AG, Ajlouni S (2017) Probiotic Delivery through Fermentation: Dairy vs. Non-Dairy Beverages. Fermentation 3:67.
View ArticlePennacchia C, Ercolini D, Blaiotta G, Pepe O, Mauriello G, Villani F (2004) Selection of Lactobacillus strains from fermented sausages for their potential use as probiotics. Meat Sci 67: 309-317. PMid:22061328
View Article PubMed/NCBIPennacchia C, Vaughan EE, Villani F (2006) Potential probiotic Lactobacillus strains from fermented sausages: Further investigations on their probiotic properties. Meat Sci 73:90-101. PMid:22062058
View Article PubMed/NCBIPanghala A, Janghub S, Virkara K, Gata Y, Kumara V, Chhikaraa N (2018) Potential non-dairy probiotic products - A healthy approach. Food Biosci 21:80-89.
View ArticleMontel MC, Buchin S, Mallet A, Delbes-Paus C, Vuitton DA, Desmasures N, Berthier F (2014) Traditional cheeses: rich and diverse microbiota with associated benefits. Int. J Food Microbiol 177:136-154. PMid:24642348
View Article PubMed/NCBIde Melo Pereira GV, de Oliveira Coelho B, Magalhaes Júnior AI, Thomaz-Soccol V, Soccol CR (2018) How to select a probiotic? A review and update of methods and criteria. Biotechnol. Adv 36 (8):2060-2076. PMid:30266342
View Article PubMed/NCBIEFSA (2016) Guidance on the preparation and presentation of an application for authorisation of a novel food in the context of Regulation (EU) 2015/2283. EFSA Journal 2016; 14(11):4594. doi: 10.2903/j.efsa.2016.4594
View ArticleCarminati C, Meucci A, Tidona F, Zago M, Giraffa G (2016) Multifunctional Lactic Acid Bacteria Cultures to Improve Quality and Nutritional Benefits in Dairy Products. In Advances in Food Biotechnology, First Edition. Edited by Ravishankar Rai V. John Wiley & Sons, Ltd. PMid:26097411
View Article PubMed/NCBIPithva S, Shekh S, Dave J, Vyas BRM (2014) Probiotic attributes of autochthonous Lactobacillus rhamnosus strains of human origin. Appl Biochem Biotechnol 173: 259-277. doi:10.1007/s12010-014-0839-9. PMid:24682879
View Article PubMed/NCBIZago M, Fornasari ME, Carminati D, Burns P, Suàrez V, Vinderola G, Reinheimer J, Giraffa G (2011) Characterization and probiotic potential of Lactobacillus plantarum strains isolated from cheeses. Food Microbiol 28:1033-1040. PMid:21569949
View Article PubMed/NCBIBaruzzi F, Quintieri L, Caputo L, Cocconcelli PS, Borcakli M, Owczarek L, Jasinska UT, Skapska S, Morea M (2016) Improvement of Ayran quality by the selection of autochthonous microbial cultures. Food Microbiol 60:92-103. PMid:27554150
View Article PubMed/NCBIDomingos-Lopes MFP, Stanton C, Ross PR, Dapkevicius MLE, Silva CCG (2017) Genetic diversity, safety and technological characterization of lactic acid bacteria isolated from artisanal Pico cheese. Food Microbiol 63:178-190. PMid:28040167
View Article PubMed/NCBIGiello M, La Storia A, Masucci F, Di Francia A, Ercolini D, Villani F (2107) Dynamics of bacterial communities during manufacture and ripening of traditional Caciocavallo of Castelfranco cheese in relation to cows' feeding. Food Microbiol 63:170-177. doi:10.1016/j.fm.2016.11.016. PMid:28040166
View Article PubMed/NCBILee HM, Lee Y (2008) A differential medium for lactic acid-producing bacteria in a mixed culture. Lett Appl Microbiol 46:676-681. PMid:18444977
View Article PubMed/NCBIRicciardi A, Guidone A, Ianniello RG, Cioffi S, Aponte M, Pavlidis D, Tsakalidou E, Zotta T, Parente E (2015) A survey of non-starter lactic acid bacteria in traditional cheeses: Culture dependent identification and survival to simulated gastrointestinal transit. Int Dairy J 43:42-50.
View ArticleCasaburi A, Di Martino V, Ferranti P, Picariello L, Villani F (2016) Technological properties and bacteriocins production by Lactobacillus curvatus 54M16 and its use as starter culture for fermented sausage manufacture. Food Control 59:31-45.
View ArticleGevers D, Huys G, Swings J, (2001) Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol Lett 205:31-36. doi: 10.1111/j.1574-6968.2001.tb10921.x. PMid:11728712
View Article PubMed/NCBIWeisburg WG, Barns SM, Pelletier DA, Lane DJ (1991) 16S Ribosomal DNA amplification for phylogenetic study. J Bacteriol 173:697-703. doi: 10.1128/jb.173.2.697-703.1991 PMid:1987160
View Article PubMed/NCBIVentura M, Canchaya C, Meylan V, Klaenhammer TR, Zink R (2003) Analysis, characterization, and loci of the tuf genes in lactobacillus and bifidobacterium species and their direct application for species identification. Appl Environ Microbiol 69:6908-6922. PMid:14602655
View Article PubMed/NCBIChen P, Zhang Q, Dang H, Liu X, Tian F, Zhao J, Chen Y, Zhang H, Chen W (2014) Screening for potential new probiotic based on probiotic properties and -glucosidase inhibitory activity. Food Control 35:65-72.
View ArticleMichael DR, Moss JWE, Lama Calvente D, Garaiova I, Plummer SF, Ramji DP (2016) Lactobacillus plantarum CUL66 can impact cholesterol homeostasis in Caco-2 enterocytes. Benef Microbes 7:443-451. PMid:26839071
View Article PubMed/NCBITomaro-Duchesneau C, Jones ML, Shah D, Jain P, Saha S, Prakash S (2014) Cholesterol Assimilation by Lactobacillus Probiotic Bacteria: An In Vitro Investigation. Biomed Res Int Article ID 380316. PMid:25295259
View Article PubMed/NCBIRudel LL, Morris MD (1973) Determination of cholesterol using o-phthalaldehyde. J Lipid Res 14:364-366. PMID: 14580182 36896-6
View ArticleCharteris WP, Kelly PM, Morelli L, Collins JK (1998) Antibiotic susceptibility of potentially probiotic Lactobacillus species. J Food Prot 61:1636-1643. PMid:9874341
View Article PubMed/NCBICLSI (Clinical and Laboratory Standards Institute) (2011) Performance Standards for Antimicrobial Susceptibility Testing. 21st Informational Supplement. M100-S21. Wayne, PA. ISBN 1-56238-742-1.
Muganga L, Liu X, Tian F, Zhao J, Zhang H, Chen W (2015) Screening for lactic acid bacteria based on antihyperglycaemic and probiotic potential and application in synbiotic set yoghurt. J Funct Foods 16:125-136.
View ArticleGuantario B, Zinno P, Schifano E, Roselli M, Perozzi G, Palleschi C, Uccelletti D, Devirgiliis C (2018) In vitro and in vivo selection of potentially probiotic Lactobacilli from Nocellara del Belice table olives. Front Microbiol 9:595. doi: 10.3389/fmicb.2018.00595 PMid:29643848
View Article PubMed/NCBISambuy Y, De Angelis I, Ranaldi G, Scarino ML, Stammati A, Zucco F (2005) The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol Toxicol 21:1-26. doi: 10.1007/s10565-005-0085-6 PMid:15868485
View Article PubMed/NCBIRoselli M, Finamore A, Britti MS, Mengheri E (2006) Probiotic bacteria Bifidobacterium animalis MB5 and Lactobacillus rhamnosus GG protect intestinal Caco-2 cells from the inflammation-associated response induced by enterotoxigenic Escherichia coli K88. Br J Nutr 95:1177-1184. doi: 10.1079/BJN20051681 PMid:16768842
View Article PubMed/NCBILiong MT, Shah NP (2005) Acid and bile tolerance and cholesterol removal ability of lactobacilli strains. J Dairy Sci 88:55-66. 72662-X
View ArticleWard LJH, Timmins MJ (1999) Differentiation of Lactobacillus casei, Lactobacillus paracasei and Lactobacillus rhamnosus by polymerase chain reaction. Lett Appl Microbiol 29: 90-92. doi: 10.1046/j.1365-2672.1999.00586.x. PMid:10499296
View Article PubMed/NCBIDesai AR, Shah NP, Powell IB (2006) Discrimination of dairy industry isolates of the Lactobacillus casei group. J Dairy Sci 89:3345-335. doi: 10.3168/jds.S0022-0302(06)72371-2. 72371-2
View ArticleSumeri I, Adamberg S, Uusna R, Sarand I, Paalme T (2012)Survival of cheese bacteria in a gastrointestinal tract simulator. Int Dairy J 25:36-41.
View ArticleJohansson ML, Molin G, Jeppsson B, Nobaek S, Ahrne S, Bengmark S (1993) Administration of different Lactobacillus strains in fermented oatmeal soup: in vivo colonization of human intestinal mucosa and effect on the indigenous flora. Appl Environ Microbiol 59:15-20. doi: 10.1128/AEM.59.1.15-20.1993. PMid:8439146
View Article PubMed/NCBIGuo CF, Zhang LW, Li JY, Zhang YC, Xue CH, Yi HX, Du M, Han X (2012) Screening of bile salt hydrolase-active lactic acid bacteria for potential cholesterol-lowering probiotic use. Adv Mat Res 345:139-146.
View ArticleLi C, Chen Y, Kwok L, Chen X, Yu H, Yang H, Yang J, Xue J, Sun T, Zhang H (2015) Identification of potential probiotic Lactobacillus plantarum isolates with broad-spectrum antibacterial activity. Dairy Sci Technol 95:381-392. doi: 10.1007/s13594-014-0206-1.
View ArticleSchmidt EJ, Boswell JS, Walsh JP, Schellenberg MM, Winter TW, Li C, Allman CW, Savage PV (2001) Activities of cholic and acid-derived antimicrobial agents against multidrug-resistant bacteria. J Antimicrob Chemother 47:671-674. PMid:11328782
View Article PubMed/NCBISolieri L, Bianchi A, Mottolese G, Lemmetti F, Giudici P (2014) Tailoring the probiotic potential of non-starter Lactobacillus strains from ripened Parmigiano Reggiano cheese by in vitro screening and principal component analysis. Food Microbiol 38:240-249. doi: 10.1016/j.fm.2013.10.003. PMid:24290648
View Article PubMed/NCBIHoráčková Š, Plocková M, Demnerová K (2018) Importance of microbial defence systems to bile salts and mechanisms of serum cholesterol reduction. Biotechnol Adv 36:682-690. doi: 10.1016/j.biotechadv.2017.12.005. PMid:29248683
View Article PubMed/NCBIMichael DR, Davies TS, Moss JWE, Lama Calvente D, Ramji DP, Marchesi JR, Pechlivanis A, Plummer SF, Hughes TR (2017) The anti-cholesterolaemic effect of a consortium of probiotics: An acute study in C57BL/6J mice. Sci Rep 7:2883 doi:10.1038/s41598-017-02889-5. PMid:28588193
View Article PubMed/NCBIGorenjak M, Gradišnik L, Trapečar M, Pistello M, Kozmus CP, Škorjanc D, Skok P, Langerholc T, Cencič A (2014) Improvement of lipid profile by probiotic/protective cultures: study in a non-carcinogenic small intestinal cell model. New Microbiol 37:51-64.
View ArticleHuang Y, Zheng Y (2010) The probiotic Lactobacillus acidophilus reduces cholesterol absorption through the down-regulation of Niemann-Pick C1-like 1 in Caco-2 cells. Br J Nutr 103: 473-478. doi: 10.1017/S0007114509991991. PMid:19814836
View Article PubMed/NCBIHuang Y, Wang J, Cheng Y, Zheng Y (2010) The hypocholesterolaemic effects of Lactobacillus acidophilus American type culture collection 4356 in rats are mediated by the downregulation of Niemann-Pick C1-like 1. Br J Nutr 104:807-812. doi:10.1017/S0007114510001285. PMid:20441669
View Article PubMed/NCBIHuang Y, Wu F, Wang X, Sui Y, Yang L, Wang J (2013) Characterization of Lactobacillus plantarum Lp27 isolated from Tibetan kefir grains: a potential probiotic bacterium with cholesterol-lowering effects. J Dairy Sci 96:2816-2825. PMid:23498003
View Article PubMed/NCBIAnandharaj M, Sivasankari B (2014) Isolation of potential probiotic Lactobacillus oris HMI68 from mother's milk with cholesterol-reducing property. J Biosci Bioeng 118(2):153-159. doi: 10.1016/j.jbiosc.2014.01.015. PMid:24613732
View Article PubMed/NCBIEFSA (2012) Guidance on the assessment of bacterial susceptibility to antimicrobials of human and veterinary importance. EFSA Journal 10:2740-2749. doi: 10.2903/j.efsa.2012.2740
View ArticleRadulović Z, Petrović T, Nedović V, Dimitrijević S, Mirković N, Petrušić M, Paunović D (2010) Characterization of autochthonous Lactobacillus paracasei strains on potential probiotic ability. Mljekarstvo 60:86-93.UDK: 637.344/579.86
Curragh HJ, Collins MA(1992) High levels of spontaneous drug resistance in Lactobacillus. J Appl Bacteriol73:31-36.
View ArticleGueimonde M, Sánchez B, de los Reyes-Gavilán CG, Margolles A (2013) Antibiotic resistance in probiotic bacteria. Front Microbiol 4, article 202, doi: 10.3389/fmicb.2013.00202. PMid:23882264
View Article PubMed/NCBIMonteagudo-Mera A, Rodríguez-Aparicio L, Rúa J, Martínez-Blanco H, Navasa N, García-Armesto MR, Ferrero MA (2012) In vitro evaluation of physiological probiotic properties of different lactic acid bacteria strains of dairy and human origin. J Funct Foods 4:531-541.
View ArticleZhang S, Liu L, Su Y, Li H, Sun Q, Liang X, Lv J (2011) Antioxidative activity of lactic acid bacteria in yogurt. Afr J Microbiol Res5:5194-5201.
View ArticleShokryazdan P, Jahromi MF, Liang JB, Sieo CC, Kalavathy R, Idrus Z, Ho YW (2017) In vitro assessment of bioactivities of Lactobacillus strains as potential probiotics for humans and chickens. J Food Sci 82:2734-2745. doi: 10.1111/1750-3841.13921. PMid:29023714
View Article PubMed/NCBI