Dr. Raghu H V.,
Mobile: +91-9466963599.
Email: Raghu.V@icar.gov.in
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 7 ISSUE: 1
Page No: 417-430
Dr. Raghu H V.,
Mobile: +91-9466963599.
Email: Raghu.V@icar.gov.in
Brijesh Kumar, Naresh Kumar, and Raghu Hirikyathanahalli Vishweswaraiah*
National Referral Centre, Dairy Microbiology Division, ICAR-National Dairy Research Institute, Karnal-132001
Cruz-Martin(mileidy@ibp.co.cu)
Hung YP(yuebin16@yahoo.com.tw)
Tawfik E(emantawfik@science.helwan.edu.eg)
Choupina A(albracho@ipb.pt)
Brijesh Kumar, Naresh Kumar, Raghu Vishweswaraiah, Spore germination-enzyme inhibition assay for rapid detection of Pesticide residue in milk (2022) Journal of Food Science & Technology 7(1):417-430
In current investigation an attempt was made to develop a bacterial enzyme inhibition-based assay for rapid detection of pesticides. In this regard different enzymes of Bacillus megaterium strain were assessed for their potential for biosensor development for pesticide detection. Among the targeted ten enzymes, eight enzymes namely ß-glucosidase, α-glucosidase, α-galactosidase, α-amylase, protease, alkaline phosphatase, peroxidase, and esterase were found expressed in used strain, however, expression time/enzyme activity was found varied among different enzymes. All expressed enzymes were screened for their activity inhibition by twenty-four pesticides of different groups using a microtiter plate assay. The inhibition of β-D-glucosidase, α-D-glucosidase, α-D-galactosidase, protease, peroxidase, and esterase was observed at pesticide concentrations of 200 ppm, 100 ppb, 10 ppb, 100 ppm, 100 ppm, and 10 ppb respectively for different pesticides. It was found that there is not a single pesticide that can be used as a model pesticide for the development of enzymatic inhibitions-based biosensors for another pesticide. The esterase was selected for further its inhibition potential due to its better reaction time i.e. 15 minutes. With a further optimized protocol, the esterase enzyme showed the inhibition at 1 ppb concentration of fenitrothion, monocrotophos, tetrachlorovinphos, paraoxon methyl, amisulbrom, ametoctradin, carbendazim, maneb, zineb, and asulam. The optimized enzyme inhibition assay offered an excellent sensitivity (limit of detection) of 0.1 ppb for captan pesticide.
Keywords: enzymes inhibition, pesticides detection, chromogenic assay
Pesticides are synthetic or biological agents utilized widely in agricultural production and public and livestock health to prevent or reduce harmful impacts from pests. In modern agriculture, pesticides have significantly increased productivity worldwide but the threat of their toxicity has also increased (Alengebawy et al. 2021; Yang et al. 2020; Ishaq et al. 2018; Korrapati et al. 2018). Detection methods for pesticide contamination need high sensitivity and accuracy because they may be present at trace levels (Mishra et al. 2012). Usually, pesticides detection techniques like Liquid Chromatography-Mass Spectrometry (LC-MS), Gas Chromatography-Mass Spectrometry (GC-MS), or High-Performance Liquid Chromatography-Mass Spectrometry (HPLC-MS) (Liu et al. 2012) are used. Though these methods are sensitive, efficient, and reliable, but need costly infrastructure establishment, complicated sample preparation steps, and are time-consuming and laborious. Consequently, there is much demand for the development of cheap, specific, and fast screening methods to allow high throughput analysis.
The inhibition of enzyme activity in the presence of a target analyte is a well-known concept. However, it has fundamentally been employed to identify pesticide residues followed by heavy metals and other inhibitors (Amine et al. 2006). The working principle of interference-based sensors generally exercises the measurement of enzyme activity in the presence and absence of pesticides. The activity of the enzyme decreases in the presence of pesticide residues and this drop in the activity can be further compared with the concentration of pesticides in the sample under analysis. Inhibition of enzyme activity in the presence of pesticide can be determined using the following formula:
I% = (Ao-Ai/Ao) x 100 (i)
Where I %- the percent inhibition, Ao- the activity of the enzyme in the absence of pesticide, Ai- the activity of the enzyme in presence of pesticide.
The above formula (i) is used for reversible or irreversible types of enzyme inhibition (Arduini et al., 2010). Different enzymes like esterase, tyrosinase, ascorbate oxidase, alkaline phosphatase, acid phosphatase, peroxidase, acetolactatesynthetase, aldehyde dehydrogenase have been used for chemical contaminants detection (Bravo et al. 2019; Dong et al. 2020; Liang et al. 2013; Marty et al. 1993; Mazzei et al. 1996; Nguyen and Jang 2021; Rekha et al. 2000; Seki et al. 1996; Vidal et al. 2008; Yang et al. 2018).
Bacterial cells are a simple, easy-to-use, and inexpensive source for biosensor development for various analytes (Kylilis et al. 2019, Gaudin V. 2017, Raut et al. 2012). The unique advantages of bacterial cells include the cost-effectiveness compared to pure enzyme preparation, effortless storage, and transport. The current study aimed to develop an enzyme inhibition-based assay for monitoring pesticide residues.
2.1. Materials
All the chemicals such as pesticides, chromogenic substrates, and organic solvents (LCMS grade) were purchased from Sigma Aldrich USA and Sodium hydroxide (NaOH) crystals were purchased from Himedia. Milli-Q water was obtained from a Milli-Q water purification system (Millipore, USA), a multimode plate reader (TECAN Infinite M200 PRO), and a Bacillus megaterium strain (IP status: Indian patent reg. no. 3819/DEL/2015).
2.2. Methodology
2.2.1. Revival, Maintenance, and activation of bacterial cells
The freeze-dried form of Bacillus megaterium strain (IP status: Indian patent reg. no. 3819/DEL/2015) was transferred in the tube having 5.0 mL of nutrient broth and incubated at 37°C for 24.0±2.0 hours to regain viability. Following incubation, a loopful of revived culture was streaked on a nutrient agar medium and incubated at 37˚C for 16.0±2.0 h. The purity of culture was examined microscopically by Gram and spore staining. The B. megaterium culture was maintained as glycerol stocks at -20 °C in the ultra-low deep freezer, until further use. For this, the culture was propagated in 50 mL of nutrient broth at 37 °C for 16±2.0 hours. The grown culture was subjected to centrifugation at 10,000 rpm for 10 minutes at 4 °C. Following centrifugation, supernatant (media components) was discarded and the cell pellet was taken. Pellet was reconstituted by the addition of 500 microliter (µL) sterile ultrapure water. Glycerol stocks were prepared by mixing equal volume, i.e. 500 µL, each of reconstituted pellet and sterile 40 % glycerol. The culture was always activated before further use by sub-culturing twice in nutrient broth and nutrient agar. Another set of cultures was stored at 4 °C and sub-cultured once a week.
2.2.2. Screening of B. megaterium for expression of marker enzymes
Preparation of test culture
The B. megaterium was streaked on a nutrient agar medium. A single colony of strain was transferred to 5.0 mL of Tryptone Glucose Yeast extract (TGY) broth and incubated for 24.0±2.0 h at 37˚C. The broth culture of the B. megaterium was centrifuged at 10,000 rotations per minute (rpm) for 10 min at 4 °C. Followed by washings of the pellet twice using 10 mili-molar (mM) potassium phosphate buffer (pH=6.8), to remove the broth components as supernatant. The final suspension was prepared in 10 mM potassium phosphate buffer (pH=6.8) and optical density was set at 595 nm to approximately 0.320±0.02 using a multimode plate reader. Final cell suspensions were further used for the screening of enzymes.
Screening protocol
The B. megaterium strain was screened for the expression of ten marker enzymes like Esterase, ß-glucosidase, α-glucosidase, α-galactosidase, α –amylase, Tryptophanase, Acid phosphatase, Protease, Alkaline phosphatase, Peorxidaseusing their respective chromogenic substrates like Indoxyl acetate, p-nitrophenyl- ß-D-glucopyranoside, p-nitrophenyl- α –D-glucopyranoside, p-nitrophenyl-α-D-galactopyranoside, Starch, 4-(Dimethylamino) cinnamaldehyde, p-Nitrophenyl Phosphatase, Azocasein, 5-Bromo,4-Chloro-3-indolyl-phosphatase, 3,3',5,5'-Tetramethylbenzidine. The protocol for screening of different marker enzymes were performed as per the protocol discussed as follows.
Esterase enzyme
For screening, 100 μL cell suspension of B. megaterium culture and 100 μL of the indoxyl acetate (chromogenic substrate) were taken in a micro-centrifuge tube (MCT). The tube was incubated at 37 ˚C and the enzyme activity was measured in terms of color development up to 4.0 h. The control tube was added with 100 μL chromogenic substrate and 10 mM potassium phosphate buffer (pH=6.8) and the absorbance was taken at 605 nm.
α-galactosidase, α-glucosidase, β-glucosidase, acid phosphatase, and alkaline phosphatase
The same protocol was used as that of esterase except for a p-nitrophenyl (PNP) chromogenic substrate (Table 1) at 10 mM concentration (3 mg/mL in 10mM PPB). The color change was observed and recorded at 10 minutes intervals initially for the first two hours followed by after 30 minutes intervals for up to 24 hours of final incubation time. Yellow color development in the tubes was indicative of the presence and expression of the marker enzyme, absorbance was measured at 405 nm.
Table 1. Targeted marker enzymes and their respective chromogenic substrates
|
S. No. |
Marker enzymes |
Chromogenic Substrate |
Concentration |
Solvent used |
|
1 |
Esterase |
Indoxyl acetate |
10 mM |
PPB pH 6.8 |
|
2 |
ß-glucosidase |
p-nitrophenyl- ß-D-glucopyranoside |
10 mM |
PPB pH 6.8 |
|
3 |
α-glucosidase |
p-nitrophenyl- α –D-glucopyranoside |
10 mM |
PPB pH 6.8 |
|
4 |
α-galactosidase |
p-nitrophenyl-α-D-galactopyranoside |
10 mM |
PPB pH 6.8 |
|
5 |
α –amylase |
Starch |
1% (w/v) |
Distilled water |
|
6 |
Tryptophanase |
4-(Dimethylamino)cinnamaldehyde |
10 mM |
Chloroform,Ethanol (1:1) |
|
7 |
Acid phosphatase |
p-Nitrophenyl Phosphatase |
10 mM |
Citrate pH 4.8 |
|
8 |
Protease |
Azocasein |
10 mM |
Tri base buffer pH 8.0 |
|
9 |
Alkaline phosphatase |
5-Bromo,4-Chloro-3-indolyl-phosphatase |
10 mM |
Tris HClbuffer pH 8.2 |
|
10 |
Peorxidase |
3,3',5,5'-Tetramethylbenzidine |
10 mM |
Na-citrate PO4 buffer pH 4.0 |
Amylase
For the detection of amylase enzyme activity in B. megaterium culture, the method of Xiao et al.(Xiao et al., 2006) was used with some modifications. The assay was initiated by adding 40 µL of starch (Sigma S-2630) solution (10 mg/mL) and 30 µL bacterial cells of in 0.1 M phosphate buffer at pH 7.0 to MCT tubes. After the incubation of 30 minutes of incubation at 50 °C, 20 µL of 1 M HCl was added to halt the enzymatic reaction, followed by the addition of 100 µL of iodine reagent. Following color development, 100 µL of the iodine-treated samples were transferred to a transparent flat-bottomed 96 well microplate and the absorbance at 580 nm was measured.
Tryptophenase
A sterilized test tube containing 4 mL of tryptophan broth was taken and inoculated with an overnight grown culture of B. megaterium culture followed by incubation at 37 °C for 24-28 hours. Then 0.5 ml of Kovac’s reagent was added to the broth culture and tubes were observed for the presence or absence of a cherry red color ring on the top of the tube.
Peroxidase
One hundred µL of substrate solution (7.63 M H2O2 + 0.921 mM TMB) in sodium citrate-phosphate buffer pH 4 was transferred to a microtiter plate containing bacterial culture followed by shaking of the plate for 15 minutes at room temperature under dark condition. Meanwhile, absorbance was taken at 650 nm.
Protease
The method was used as per Mel et al.(Mel et al., 2000) with some modifications. One hundred µL of azocasein (5 mg/mL) in 100 mMTris (pH 8.0) was taken in MCT followed by the addition of 100 µL of cell suspension of B. megaterium and the mixture was incubated at 37 °C for 1 hour. Then the enzyme reaction was stopped by the addition of 400 µL of 10% trichloroacetic acid (in Mili Q water). The final mixture was centrifuged at 10,000 rpm for 10.minutes and a supernatant of trichloroacetic acid was transferred in 700 µL of 525 mMNaOH. The optical density (O.D) of the final mixture was measured at 440 nm.
2.2.3. Screening of marker enzyme for pesticide inhibition
Targeted Pesticides
Twenty-four pesticides (Fenitrothion, Monocrotophos, Tetrachlorovinphos, Malathion, Dimethoate, Paraoxon methyl, Endosulfan, Aldrin, Amisulbrom, Ametoctradin, Edinfenphos
Carbendazim, Maneb, Zineb, Captan, Thiram, Ziram, Glyphosate, 2-Phenylphenol, Thiobencarb, Alachlor, Atrazine, Asulam, and Aclonifen) were analyzed for their inhibition activity against targeted enzymes. The procedure for the screening of marker enzyme for its inhibition study used is as follows.
Step 1. Exposure: A 50 μL of activated bacterial cells (OD600 0.320±0.02) were transferred to wells of a microtiter plate containing different concentrations of pesticide/solvent residues. Following proper mixing, the plate was allowed to incubate at 37°C for 1 hour.
Step 2. Enzyme-substrate reaction: A 50 μL of chromogenic substrate was added to each well of the microtiter plate. After the addition of the substrate, the tubes were allowed to incubate at 37 °C for an enzyme-substrate reaction to take place. The optical density was taken at specific intervals of time using a multimode plate reader. Final observations for inhibition in enzyme activity were taken in terms of color development after the optimum incubation period depending upon the type of enzyme to be screened for inhibition.
2.2.4. Optimization of enzyme inhibition assay with a selected enzyme
Quantity of bacterial cells
10, 20, 30, 40, and 50 µL of activated bacterial cells (Optical density 0.320±0.02) of B. megaterium strain reconstitution with phosphate buffer was used. The enzyme reaction was observed followed by adding 50 µL (10 mM) of the respective chromogenic substrate.
2.2.5. Incubation time
Time of exposure
The activity of the selected enzyme was allowed to inhibit by exposure to a minimum concentration of pesticide for a different period ranging from 10-50 minutes.
Enzyme-substrate reaction time
After exposure with pesticide for optimized time, 50 µL(10 mM) of the chromogenic substrate was added in the wells of the microtiter plate followed by incubation for the detection of residual enzyme activity from 5 to 35 minutes.
3.1. Enzyme (s) expression
The Bacillus megaterium (B. megaterium) strain was screened for α-amylase activity as depicted in Figure 1A. A significant level of amylase, α-galactosidase, ß-glucosidase, alkaline phosphatase, α-glucosidase, α –amylase, Protease, Peroxidase, and Esterase was found at 70, 35, 30, 40, 65, 60, 15, and 15 min, respectively (Figure 1A and 1B). Whereas in case of acid phosphatase and tryptophenase enzyme activity, B. megaterium has not shown any enzyme activity even at or above 120 min incubation at 37 oC. In the existing literature, almost all species of the Bacillus genus are known to synthesize α-amylase, since this genus has the potential to dominate the enzyme industry (Vishnu et al. 2014). In another study, Gurudeeban et al. (Gurudeeban et al. 2011) have isolated a B. megaterium from the leaves of Avicennia marina able to express amylase. Stark et al. (Stark et al. 1982) who also reported the production of the α-amylase enzyme in a B. megaterium strain S218. However, this enzyme was found intracellular in B. megaterium strain M as shown by Weibull et al. (Weibull et al. 1959). Thus acid phosphatase may not be released extracellularly to show its activity. Our results are well supported by Patil et al. (A. G. G. Patil et al. 2010) who also reported the expression of α- galactosidase in B. megaterium VHM1. This α-glucosidase enzyme is well reported for its widespread occurrence in several species of the Bacillus (Castro et al. 1995). Our results in this regard were well supported by Kelly and Fogarty (Kelly and Fogarty 1983) who also reported the expression of α-glucosidase in B. megaterium. In another study by Stark et al. (Stark et al. 1982) also reported the expression of the α-glucosidase enzyme in B. megaterium S218.The production of β-D-glucosidases by several Bacillus species such as B. subtilis, B. licheniformis, etc. is well reported in the literature (Bagudo et al. 2014; Naz et al. 2010; Rehena et al. 1989). Castro et al. (Castro et al. 1995)who found that the presence of this enzyme extracellularly as well as membrane-bound. Higerd and Spizizen(1973) who also found expression of the same enzyme in vegetative cells of many other Bacillus species. Jung et al. (2003) also reported the presence of the same enzyme in cell-bound in the B. megaterium strain. Similarly, Priest (1977) also observed the expression of the same enzyme in the form of extracellular enzymes in the genus Bacillus including B. megaterium and B. licheniformis. Zheng et al. (2017) also reported the activity of the same enzyme B. megaterium who studied stereoselectivity and catalytic activity of this enzyme.Tariq et al. (2016) who also reported negative tryptophanase activity in B. megaterium. Our finding in this regard was well supported by Priya et al. (2014) and Wood and Tristram(1970) they also reported the production of alkaline phosphatase by several Bacillus species such as B. megaterium, B. subtilis, etc. Our study in this regard was well supported by findings of other researchers they also reported production of protease by several Bacillus species such as B. megaterium, B. subtilis, etc. The production of peroxidase by several Bacillus species such as B. megaterium, B. subtilis, etc. (Patil 2014; Rao andKavya 2014).Further, all expressed enzymes in the B. megaterium were subsequently screened for inhibition by all targeted pesticides of different groups.
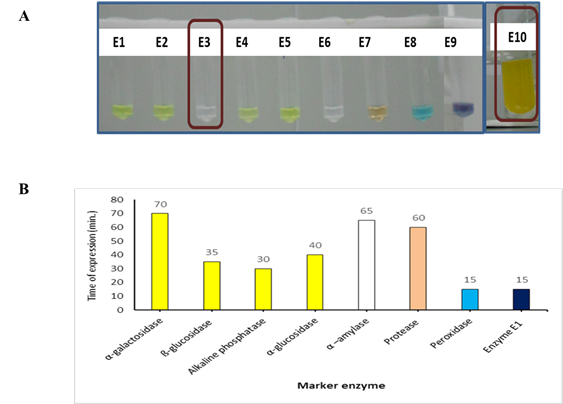
Figure 1. Enzyme expression in Bacillus megaterium. A. Enzyme expression in Bacillus megaterium using specific chromogenic substrate. B. Time of expression (or change in color of the chromogenic substrate) of enzymes of Bacillus megaterium strain
3.2. Screening enzymes for their inhibition by pesticides
Results of inhibition of α-glucosidase, ß-glucosidase, α-galactosidase, and esterase enzyme activity by different pesticide screened by microtiter assay are shown in Table 2-5 and summarized limit of detection of enzymes are given in Table 6. Based on visual observation, inhibition of α-glucosidase enzyme activity was found up to 100 ppb with maneb followed by thiram and ziram up to 1 ppm in terms of no yellow color i.e., p-nitrophenol development due to the inability of the α-glucosidaseenzyme to hydrolyze PNP substrate. Other twenty-two pesticides showed inhibition up to a maximum of 1 ppm only. In case of ß-glucosidase enzyme, inhibition of activity was found up to 200 ppm with asulam and followed by glyphosate up to 300 ppm with ß-glucosidase indicated by no p-nitrophenol production or yellow color development. Other pesticides were not inhibitory to ß-glucosidaseactivity up to the highest concentration of 500 ppm. Further, α-galactosidase was sensitive to most pesticides and more interestingly, pesticides that showed the highest inhibition were fungicides. Based on visual observation, the highest inhibition was observed up to 10 ppb with maneb followed by edinfenphos, thiram, ziram up to 1 ppm. Some pesticides were not inhibitory to enzyme activity up to 100 ppm. The alkaline phosphatase activity of was not inhibited by any of the pesticides in terms of yellow color i.e., p-nitrophenol development due to the enzymatic hydrolysis of PNP substrate. The activity of α-amylase was also not inhibited by any of the pesticides in terms of no blue color development due to the enzymatic hydrolysis of starch. The inhibition of protease enzyme activity was observed only with zineb at 100 ppm in terms of lesser red color development due to the partial ability of the enzyme to utilize azo-casein. The inhibition of enzyme activity was observed only with ziram at 100 ppm in terms of lesser blue color development due to the partial ability inhibition of the enzyme activity. Later, esterase enzyme activity studies wherein no change in color of indoxyl acetate from pink to purple in the presence of pesticides may be due to inhibition by 10 ppb for fenitrothion, monocrotophos, malathion, paraoxon methyl amisulbrom, ametoctradin, carbendazim, maneb, captan, asulum was observed. Our findings in this regard were very well supported by Ahmed et al. (Ahmed et al. 2002) who also reported the significant role of protease for insecticide resistance in a strain of Muscadomestica L. Our results in this regard are very well supported by Niemi et al. (2009). Similarly, another study was done by Bhardwaj and Shekhar(Bhardwaj and Shekhar 2005) who also reported inhibition of α-glucosidaseand α-galactosidaseactivity in the midgut of the last instar naiad of Trithemis aurora by quinalphos, chlorpyrifos, cypermethrin, and deltamethrin pesticides. Inhibition in the ß-glucosidase enzymatic activity in tea garden soil containing residues of organophosphate and organochlorine pesticides were shown by Bishnu et al. (2008). A significant role of protease for insecticide resistance in a strain of Muscadomestica L was reported by Ahmed et al. (2002). Moccelini et al. (2010)developed a biosensor based on the inhibition of peroxidase for the detection of thiodicarb and carbamate pesticides. Using microtiter assay, among eight enzymes screened for their inhibition by twenty-four pesticides, only seven enzymes namely esterase, β-glucosidase, α-glucosidase, α-galactosidase, α-amylase, protease, alkaline phosphatase, and peroxidase showed sensitivity towards pesticides. However, a higher level of sensitivity in terms of lower LOD, i.e., 10 ppb for pesticides was obtained with α-galactosidase and enzyme esterase but enzyme esterase has shown a lesser time i.e. 15 minutes for enzyme reaction as compared to other enzymes. Therefore, esterase enzyme activity in Bacillus species was selected for further optimization study.
Table 2. Visual perception based inhibition of α-glucosidase activity
|
S. No. |
Pesticide |
LOD (ppm) |
S. No. |
Pesticide |
LOD (ppm) |
|
1 |
Fenitrothion |
>100 |
13 |
Maneb |
0.1 |
|
2 |
Monocrotophos |
>100 |
14 |
Zineb |
100 |
|
3 |
Tetrachlorovinphos |
>100 |
15 |
Captan |
10 |
|
4 |
Malathion |
>100 |
16 |
Thiram |
1 |
|
5 |
Dimethoate |
>100 |
17 |
Ziram |
1 |
|
6 |
Paraoxon methyl |
>100 |
18 |
Glyphosate |
>100 |
|
7 |
Endosulfan |
100 |
19 |
2-Phenylphenol |
>100 |
|
8 |
Aldrin |
>100 |
20 |
Thiobencarb |
>100 |
|
9 |
Amisulbrom |
>100 |
21 |
Alachlor |
>100 |
|
10 |
Ametoctradin |
>100 |
22 |
Atrazine |
>100 |
|
11 |
Edinfenphos |
>100 |
23 |
Asulam |
100 |
|
12 |
Carbendazim |
>100 |
24 |
Aclonifen |
>100 |
Table 3. Visual perception based inhibition of ß-glucosidase activity
|
S. No. |
Pesticide |
LOD (ppm) |
S. No. |
Pesticide |
LOD (ppm) |
|
1 |
Fenitrothion |
>500 |
13 |
Maneb |
>500 |
|
2 |
Monocrotophos |
>500 |
14 |
Zineb |
>500 |
|
3 |
Tetrachlorovinphos |
>500 |
15 |
Captan |
>500 |
|
4 |
Malathion |
>500 |
16 |
Thiram |
>500 |
|
5 |
Dimethoate |
>500 |
17 |
Ziram |
>500 |
|
6 |
Paraoxon methyl |
>500 |
18 |
Glyphosate |
300 |
|
7 |
Endosulfan |
>500 |
19 |
2-Phenylphenol |
>500 |
|
8 |
Aldrin |
>500 |
20 |
Thiobencarb |
>500 |
|
9 |
Amisulbrom |
>500 |
21 |
Alachlor |
>500 |
|
10 |
Ametoctradin |
>500 |
22 |
Atrazine |
>500 |
|
11 |
Edinfenphos |
>500 |
23 |
Asulam |
200 |
|
12 |
Carbendazim |
>500 |
24 |
Aclonifen |
>500 |
Table 4. Visual perception based inhibition of α-galactosidase activity
|
S. No. |
Pesticide |
LOD (ppm) |
S. No. |
Pesticide |
LOD (ppm) |
|
1 |
Fenitrothion |
100 |
13 |
Maneb |
0.01 |
|
2 |
Monocrotophos |
100 |
14 |
Zineb |
10 |
|
3 |
Tetrachlorovinphos |
100 |
15 |
Captan |
10 |
|
4 |
Malathion |
>100 |
16 |
Thiram |
1 |
|
5 |
Dimethoate |
>100 |
17 |
Ziram |
1 |
|
6 |
Paraoxon methyl |
>100 |
18 |
Glyphosate |
>100 |
|
7 |
Endosulfan |
100 |
19 |
2-Phenylphenol |
>100 |
|
8 |
Aldrin |
>100 |
20 |
Thiobencarb |
100 |
|
9 |
Amisulbrom |
>100 |
21 |
Alachlor |
100 |
|
10 |
Ametoctradin |
>100 |
22 |
Atrazine |
>100 |
|
11 |
Edinfenphos |
1 |
23 |
Asulam |
100 |
|
12 |
Carbendazim |
>100 |
24 |
Aclonifen |
>100 |
Table 5. Visual perception based inhibition for esterase activity
|
S. No. |
Pesticide |
LOD (ppm) |
S. No. |
Pesticide |
LOD (ppm) |
|
1 |
Fenitrothion |
0.01 |
13 |
Maneb |
0.01 |
|
2 |
Monocrotophos |
0.01 |
14 |
Zineb |
0.1 |
|
3 |
Tetrachlorovinphos |
0.01 |
15 |
Captan |
0.01 |
|
4 |
Malathion |
0.1 |
16 |
Thiram |
0.1 |
|
5 |
Dimethoate |
0.1 |
17 |
Ziram |
0.1 |
|
6 |
Paraoxon methyl |
0.01 |
18 |
Glyphosate |
10 |
|
7 |
Endosulfan |
10 |
19 |
2-Phenylphenol |
0.1 |
|
8 |
Aldrin |
1 |
20 |
Thiobencarb |
50 |
|
9 |
Amisulbrom |
0.01 |
21 |
Alachlor |
50 |
|
10 |
Ametoctradin |
0.01 |
22 |
Atrazine |
10 |
|
11 |
Edinfenphos |
0.1 |
23 |
Asulam |
0.01 |
|
12 |
Carbendazim |
0.01 |
24 |
Aclonifen |
10 |
Table 6. Summary of observations on maker enzymes screening using microtiter assay
|
S. No. |
Marker enzymes |
Sensitivity |
Pesticide |
Reaction time |
|
1 |
α-glucosidase |
100 ppb |
Thiram, Ziram, Maneb |
2.50 hour |
|
2 |
ß-glucosidase |
200 ppm |
Asulam |
35 minutes |
|
3 |
Acid phosphatase |
- |
- |
- |
|
4 |
α-galactosidase |
10 ppb |
Maneb |
5.50 hour |
|
5 |
Alkaline phosphatase |
>100 ppm |
- |
30 minutes |
|
6 |
α –amylase |
>100 ppm |
- |
2 hours |
|
7 |
Protease |
100 ppm |
Zineb |
1 hour |
|
8 |
Peorxidase |
100 ppm |
Ziram |
15 minutes |
|
9 |
Esterase |
10 ppb |
Captan, Maneb, Asulam etc. |
15 minutes |
|
10 |
Tryptophanase |
- |
- |
- |
3.3. Optimization of colorimetric microtiter assay with an esterase enzyme
As shown in Figure 2A, the activity of esterase at 30 µL of spores was observed with optimum color development within a short period of incubation, i.e., 10 minutes. Exposure is the pre-incubation of the enzyme with pesticide residues before the addition of substrate. In the current work, the activity of esterase was allowed to inhibit by exposure to 10 ppb of asulam for a different period ranging from 10-50 minutes. As shown in Figure 2B, an increasing trend in the degree of the inhibitory signal from 12.54 to 32.98 % at 10 ppb as the time of exposure was increased from 5 min. to 40 min. An increasing trend in inhibition of esterase activity was found to increase from 20.50 to 45.11 % at 10 ppb with an increase in time of incubation from 5 minutes to 35 minutes (Figure 2C). After that, no significant increase in inhibition of esterase activity was observed at 30 minutes of incubation. However, at 40 minutes of incubation, the activity of esterase was increased and masked inhibition. Thus by these findings, an incubation time of 10 minutes was selected for enzyme-substrate reaction to take place. Our findings are similar to the outcomes of Bucur et al. (Bucur et al. 2006) who observed an increase in inhibition of AChE by carbaryl with the simultaneous increase in exposure time.
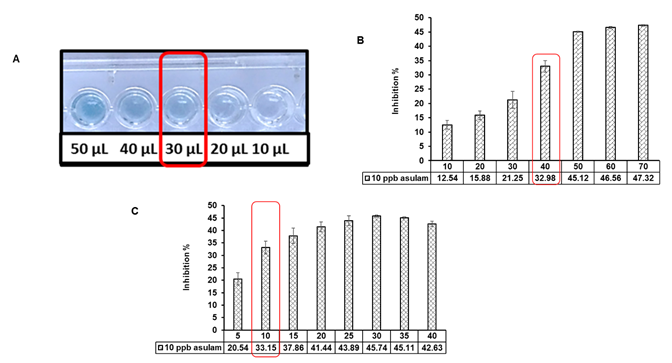
Figure 2. Optimization of chromogenic microtiter assay for detection of pesticide residues in water. A. Volume of spores B. Time of exposure (minutes) C. Time of reaction (minutes)
3.4. Determination of the limit of detection (LOD)
The LOD of each pesticide by developed assay was taken as the pesticide concentration that produced blue color for microtiter plate as well as an inhibition at ≥ 30 % in esterase activity with an O.D. at 605 nm was measured in microtiter plate assay to avoid any false-positive results. LOD obtained for 24 pesticides of insecticide, herbicide, and fungicide group with microtiter assay are summarized in Table 7. With the optimized protocol, the esterase enzyme showed the most significant inhibition (i.e. limit of detection) at a with a further optimized protocol, the esterase enzyme showed the inhibition at 1 ppb concentration of fenitrothion, monocrotophos, tetrachlorovinphos, paraoxon methyl, amisulbrom, ametoctradin, carbendazim, maneb, zineb, and asulam in a reaction time of 10 minutes. Most significantly a LOD of 0.1 ppb for captan pesticide was achieved in a reaction time of 15 minutes. The percent inhibition in esterase activity was found to increase with a simultaneous increase in the concentration of pesticide which is in line with the findings of other studies (Ayat et al. 2021; Yunhe et al. 2010). Comparing the analytical data obtained for pesticides of three groups indicated the lower LOD and thus higher sensitivity for members of fungicides followed by insecticides.
Table 7. LOD of different groups of pesticides achieved using microtiter assay
|
S. No. |
Pesticide Name |
LOD |
S. No. |
Pesticide Name |
LOD |
|
1.
|
Fenitrothion |
1 ppb |
13. |
Maneb |
1 ppb |
|
2. |
Monocrotophos |
1 ppb |
14. |
Zineb |
1 ppb |
|
3. |
Tetrachlorvinphos |
1 ppb |
15. |
Captan |
0.1 ppb |
|
4. |
Malathion |
10 ppb |
16. |
Thiram |
10 ppb |
|
5. |
Dimethoate |
10 ppb |
17. |
Ziram |
10 ppb |
|
6. |
Paraoxon-methyl |
1 ppb |
18. |
Glyphosate |
5 ppm |
|
7. |
Endosulfan |
5 ppm |
19. |
O-phenylphenol |
10 ppb |
|
8. |
Aldrin |
750 ppb |
20. |
Thiobencarb |
10 ppm |
|
9. |
Amisulbrom |
1 ppb |
21. |
Alachlor |
10 ppm |
|
10. |
Ametoctradin |
1 ppb |
22. |
Atrazine |
10 ppm |
|
11. |
Edifenphos |
10 ppb |
23. |
Asulam |
1 ppb |
|
12. |
Carbendazim |
1 ppb |
24. |
Aclonifen |
10 ppm |
For insecticide detection, several biosensors are developed and achieved the different limits of detection. The higher sensitivity of some of the organophosphate pesticides can be explained by the fact that these pesticides cause inhibition by forming stable covalent intermediates (Kumaran and Tran-Minh 1992; Sharma et al. 2021; Wu et al. 2021). Montes et al. (Montes et al. 2018) developed an electrochemical biosensor based on optimized biocomposite for organophosphorus and carbamates achieved LOD of 0.25 ± 0.03 to 1.03 ± 0.05 ppb for malathion. Gan et al. (Gan et al., 2010) developed a disposable organophosphorus pesticides (OPs) enzyme biosensor based on magnetic composite nanoparticle-modified screen-printed carbon electrodes (SPCE) and detected dimethoate up to the lowest limit of 56 ppb. Gabaldónet al. (Gabaldón et al. 1999) reported a commercial kit available for fenitrothion detection with a LOD of 25 ppb. Wu et al. (Wu et al. 2011) developed amperometricacetylcholinesterase (AChE) biosensor which was fabricated based on mesocellular silica foam achieved LOD 0.05 ppb for monocrotophos. For fungicides, Marty et al. (Marty et al., 1993) developed a sensor based on the enzyme aldehyde dehydrogenase for detection of dithiocarbamate fungicide, i.e., maneb, and reported LOD of 0.05 ppm. An enzyme aldehyde dehydrogenase-based biosensor for the detection of zineb was developed with a LOD of 8 ppb (Noguer et al., 1999). Pita et al. (Pita et al. 1997) developed a biosensor based on enzyme tyrosinase for the detection of ziram and reported a LOD of 22.63 ppb with the developed sensor. The least sensitivity was achieved for the herbicides group at 10 ppm for some of its members. This shows that these pesticides are low inhibitory to esterase. For herbicide detection, several biosensors are developed and achieved different LOD, but most of the sensors are based on other than esterase. For example, Koblizek et al.(Koblizek et al. 1998) developed a biosensor based on photosystem-II (PSII) particles from Synechococcus elongates for detection of atrazine and got LOD up to 0.43 ppb. Oliveira et al. (Oliveira et al. 2012) developed a biosensor based on heme-containing enzymes and achieved a LOD of 30 ppb for glyphosate. An immunoassay (ELISA) is commercially available by Abraxis LLC for glyphosate, atrazine, and alachlor with LOD of 0.05 ppb, 3 ppb, and 0.08 ppb respectively (Abraxis LLC).The pesticide paraoxon is the strong known organophosphate inhibitor for the activity of various enzymes and was used as a model pesticide in the development of several enzymatic inhibitions based biosensors in existing prior-art (Arduini et al. 2007; Heo and Crooks 2005; Ivanov et al. 2012; Mulchandani et al. 2006; Pohanka et al. 2010). Our findings show that paraoxon does not represent the inhibition pattern of other pesticides against enzymes. That infers that paraoxon methyl is not a model pesticide for the development of enzymatic inhibitions-based biosensors for another pesticide.
All expressed enzymes were screened for their activity inhibition by twenty-four pesticides of different groups using the enzyme inhibition assay. The limit of detection of β-D-glucosidase, α-D-glucosidase, α-D-galactosidase, protease, peroxidase, and esterase was observed at pesticide concentrations of 200 ppm, 100 ppb, 10 ppb, 100 ppm, 100 ppm, and 10 ppb respectively for different pesticides. With the optimized esterase inhibition assay, a LOD of 0.1 ppb for captan pesticide was achieved in a reaction time of 10 minutes. The findings of the current study are consistent with the existing studies indicating the potential of esterase to target the detection of a broad range of pesticides. The enzyme inhibition-based assay using esterase enzyme would be a promising approach for the rapid and cost-effective assay for pesticide detection.
The authors are thankful to the National Referral Centre for Milk Quality and Safety of the ICAR-NDRI for providing facilities for research and the University Grant Commission (UGC) for granting research fellowships.
Disclosure statement
The authors solely conducted the study design, execution, interpretation, and manuscript preparation.
Funding
The authors thank the University Grant Commission (UGC) for granting research fellowships.
Ahmed S, Wilkins RM, Mantle D (2002) Comparative effect of various insecticides on intracellular proteases in an insecticide-resistant and susceptible strain of Muscadomestica L. Online J. Biolog., 2:183-185.
View ArticleAlengebawy, A., Abdelkhalek, S. T., Qureshi, S. R., & Wang, M. Q. (2021). Heavy Metals and Pesticides Toxicity in Agricultural Soil and Plants: Ecological Risks and Human Health Implications. Toxics, 9(3), 42. PMid:33668829
View Article PubMed/NCBIAmine A, Mohammadi H, Bourais I, Palleschi G (2006) Enzyme inhibition-based biosensors for food safety and environmental monitoring. Biosens.Bioelectron.,21: 1405-1423. PMid:16125923
View Article PubMed/NCBIArduini F, Amine A, Moscone D, Palleschi G (2010). Biosensors based on cholinesterase inhibition for insecticides, nerve agents and aflatoxin B1 detection (review. Microchimica Acta, 170: 193-214.
View ArticleArduini F, Amine A, Moscone D, Ricci F, Palleschi G (2007). Fast, sensitive and cost-effective detection of nerve agents in the gas phase using a portable instrument and an electrochemical biosensor. Anal. Bioanalytic. Chem., 388(5-6): 1049-1057. PMid:17508205
View Article PubMed/NCBIAsker MM, Mahmoud MG, El Shebwy K, Aziz MSA (2013). Purification and characterization of two thermostable protease fractions from Bacillus megaterium. J. Gen. Eng.Biotechnol.,11(2): 103-109.
View ArticleAyat M, Ayouz K, Yaddadene C, Berouaken M, Gabouze N (2021). Porous silicon-modified electrode for electrochemical pesticide biosensor. J. Coat. Tech. Res., 18(1): 53-62.
View ArticleBagudo AI, Argungu AU, Aliero AAS, Suleiman N, Kalpana S (2014). Bacillus subtilis as an alternative source of beta-glucosidase. Int J Mod Cell Mol Biol, 3(1): 1-9.
Bhardwaj AC, Shekhar C (2005). Pesticides eclyptic influence on activity of midgutcarbohydrases of Trithemis aurora (Burm.)(Anisptera: Libellulidae) naiads. J. Appl. Zoolog. Res.,16(1): 117-118.
Bishnu A, Saha T, Mazumdar D, Chakrabarti K, Chakraborty A (2008). Assessment of the impact of pesticide residues on microbiological and biochemical parameters of tea garden soils in India. J. Environ. Sci. Health Part B, 43(8): 723-731. PMid:18941998
View Article PubMed/NCBIBravo I, Gutiérrez-Sánchez C, García-Mendiola T, Revenga-Parra M, Pariente F, Lorenzo E (2019) Enhanced Performance of Reagent-Less Carbon Nanodots Based Enzyme Electrochem. Biosen.Sen.,19(24): 5576-5576. PMid:31861148
View Article PubMed/NCBIBucur B, Fournier D, Danet A, Marty JL (2006) Biosensors based on highly sensitive acetylcholinesterases for enhanced carbamate insecticides detection. Analy. Chemic. Acta, 562(1): 115-121.
View ArticleCastro GR, Baigori MD, Sineriz F (1995) Production of α-glucosidase by Bacillus sp. Strains. Acta Biotechnol.,15(2): 233-240.
View ArticleDong J, Yan, H, Li Y, Liu A, Wei W, Liu S (2020) Fluorescence sensor for organophosphorus pesticide detection based on the alkaline phosphatase-triggered reaction. Analyt. Chemica Acta, 102-108. PMid:32928470
View Article PubMed/NCBIGabaldón JA, Maquieira A, Puchades R (1999). Current trends in immunoassay-based kits for pesticide analysis. Crit. Rev. Food Sci. Nutri., 39(6): 519-538. PMid:10595298
View Article PubMed/NCBIGan N, Yang X, Xie D, Wu Y, Wen W (2010). A disposable organophosphorus pesticides enzyme biosensor based on magnetic composite nano-particles modified screen printed carbon electrode. Sen., 10(1): 625-638. PMid:22315558
View Article PubMed/NCBIGaudin V. (2017). Advances in biosensor development for the screening of antibiotic residues in food products of animal origin - A comprehensive review. Biosensors & bioelectronics, 90, 363-377. PMid:27940240
View Article PubMed/NCBIGurudeeban S, Satyavaniand K, Ramanathan T (2011). Production of extra cellular-amylase using Bacillus megaterium isolated from White Mangrove (Avicennia marina). Asian J. Biotechnol., 3(3): 310-316.
View ArticleHackett RH, Setlow P (1983). Enzymatic activity of precursors of Bacillus megaterium spore protease. J. Bacteriol., 153(1): 375-378. PMid:6401283
View Article PubMed/NCBIHeo J, Crooks RM (2005). Microfluidic biosensor based on an array of hydrogel-entrapped enzymes. Anal. Chem., 77(21): 6843-6851. PMid:16255581
View Article PubMed/NCBIHigerd TB, Spizizen J (1973). Isolation of two acetyl esterases from extracts of Bacillus subtilis. J. Bacteriol., 114(3): 1184-1192. PMid:4197268
View Article PubMed/NCBIIshaq Z, Sajid MW, Saleem S, Mehmood A, Ali L, Hussain A (2018). A perspective on organochlorine pesticide residues in milk produced in Pakistan. EC Nutrit., 13(6): 402-410.
IvanovY, Marinov I, Portaccio M, Lepore M, Mita DG, Godjevargova T (2012). Flow-injection system with site-specific immobilization of acetylcholinesterase biosensor for amperometric detection of organophosphate pesticides. Biotechnol. Equip., 26(3): 3044-3053.
View ArticleJung YJ, Lee JK, Sung CG, Oh TK, Kim HK (2003). Nonionic detergent-induced activation of an esterase from Bacillus megaterium 20-1. J.Mol. Cat. B: Enz.,26(3-6): 223-229.
View ArticleKelly CT, Fogarty WM (1983). Microbial α-glucosidases. Process Biochem.,18: 6-12.
Koblizek M, Masojidek J, Komenda J, Kucera T, Pilloton R, Mattoo AK, Giardi MT (1998). A sensitive photosystem-II based biosensor for detection of a class of herbicides. Biotechnol.Bioeng.,60(6): 664-669. 1097-0290(19981220)60:6<664::AID-BIT3>3.0.CO;2-B
View ArticleKorrapati K, Kotha K, Nelapati K (2018). Determination of organophosphorus pesticide residues in fodder samples along Musi river belt, Hyderabad, India. Int. J. Curr. Microbiol. App. Sci, 7(4): 2535-2545.
View ArticleKumaran S, Tran-Minh (1992). Determination of organophosphorous and carbomate insecticides by flow injection analysis. Anal. Biochem, 200: 187-194. 90297-K
View ArticleKylilis, N., Riangrungroj, P., Lai, H. E., Salema, V., Fernández, L. Á., Stan, G. B. V., ... & Polizzi, K. M. (2019). Whole-cell biosensor with tunable limit of detection enables low-cost agglutination assays for medical diagnostic applications. ACS sensors, 4(2), 370-378. PMid:30623662
View Article PubMed/NCBILiang M, Fan K, Pan Y, Jiang H, Wang F, Yang D, Yan X (2013). Fe3O4 magnetic nanoparticle peroxidase mimetic-based colorimetric assay for the rapid detection of organophosphorus pesticide and nerve agent. Analyt. Chem., 85(1): 308-312. PMid:23153113
View Article PubMed/NCBILiu H, Xiang Y, Lu Y, Crooks RM (2012). Aptamer‐based origami paper analytical device for electrochemical detection of adenosine. Angewandte Chemie, 124(28): 7031-7034.
View ArticleMarty JL, Mionetto N, Noguer T, Ortega F, Roux C (1993). Enzyme sensors for the detection of pesticides. Biosen.Bioelectron.,8(6): 273-280. 85007-B
View ArticleMazzei, F., Botrè, F., & Botrè, C. (1996). Acid phosphatase/glucose oxidase-based biosensors for the determination of pesticides. Analytica Chimica Acta, 336(1-3), 67-75. 00378-9
View ArticleMedzon, E. L., & Brady, M. L. (1969). Direct measurement of acetylesterase in living Protist cells. Journal of Bacteriology, 97(1), 402-415. PMid:4974398
View Article PubMed/NCBIMel, S. F., Fullner, K. J., Wimer-Mackin, S., Lencer, W. I., & Mekalanos, J. J. (2000). Association of protease activity in Vibrio cholera vaccine strains with decreases in transcellular epithelial resistance of polarized T84 intestinal epithelial cells. Infection and Immunity, 68(11), 6487-6492. PMid:11035765
View Article PubMed/NCBIMishra, R. K., Dominguez, R. B., Bhand, S., Muñoz, R., & Marty, J. L. (2012). A novel automated flow-based biosensor for the determination of organophosphate pesticides in milk. Biosensors and Bioelectronics, 32(1), 56-61. PMid:22221795
View Article PubMed/NCBIMoccelini, S. K., Vieira, I. C., De Lima, F., Lucca, B., Barbosa, A. M. J., & Ferreira, V. S. (2010). Determination of thiodicarb using a biosensor based peroxidase immobilized on alfalfa sprout in self-assembled monolayers. Talanta, 82(1), 164-170. PMid:20685452
View Article PubMed/NCBIMontes, R., Céspedes, F., Gabriel, D., & Baeza, M. (2018). Electrochemical biosensor based on optimized biocomposite for organophosphorus and carbamates pesticides detection. Journal of Nanomaterials. 2018, 1-13
View ArticleMulchandani, P., Chen, W., & Mulchandani, A. (2006). Microbial biosensor for direct determination of nitrophenyl-substituted organophosphate nerve agents using genetically engineered Moraxella sp. AnalyticaChimica, Acta,568, 217-221. PMid:17761263
View Article PubMed/NCBINaz, S., Ikram, N., Rajoka, M. I., Sadaf, S., & Akhtar, M. W. (2010). Enhanced production and characterization of a β-glucosidase from Bacillus halodurans expressed in Escherichia coli.Biochemistry (Moscow), 75(4), 513-518. PMid:20618142
View Article PubMed/NCBINguyen, D. K., & Jang, C. H. (2021). An acetylcholinesterase-based biosensor for the detection of pesticides using liquid crystals confined in microcapillaries. Colloids and Surfaces B: Biointerfaces, 200, 111587-111587. PMid:33529929
View Article PubMed/NCBINiemi, R. M., Heiskanen, I., Ahtiainen, J. H., Rahkonen, A., Mäntykoski, K., Welling, L., Laitinen, P., & Ruuttunen, P. (2009). Microbial toxicity and impacts on soil enzyme activities of pesticides used in potato cultivation. Applied Soil Ecology, 41(3), 293-304.
View ArticleNoguer, T., Gradinaru, A., Ciucu, A., & Marty, J. L. (1999). A new disposable biosensor for the accurate and sensitive detection of ethylenebis (dithiocarbamate) Fungicides. Fungicides.Analytical Letters, 32(9), 1723-1738.
View ArticleOliveira, G. C., Moccelini, S. K., Castilho, M., Terezo, A. J., Possavatz, J., Magalhães, M. R., & Dores, E. F. (2012). Biosensor based on atemoya peroxidase immobilised on modified nanoclay for glyphosate biomonitoring. Talanta, 98, 130-136. PMid:22939138
View Article PubMed/NCBIPadmapriya, M., & Williams, B. C. (2017). Purification and characterization of neutral protease enzyme from Bacillus subtilis. Journal of Microbiology and Biotechnology Research, 2(4), 612-618.
Pant, G., Prakash, A., Pavani, J. V. P., Bera, S., Deviram, G. V. N. S., Kumar, A., Panchpuri, M., & Prasuna, R. G. (2015). Production, optimization and partial purification of protease from Bacillus subtilis. Journal of Taibah University for Science, 9(1), 50-55.
View ArticlePatil, A. G. G., Kumar, P., Veerappa H., M., Yaligara, V., & Kyoung, L. (2010). α-galactosidase from Bacillus megaterium VHM1 and its application in removal of flatulence-causing factors from soymilk. Journal of Microbiology and Biotechnology, 20, 1546-1554. PMid:21124061
View Article PubMed/NCBIPatil, S. R. (2014). Production and purification of lignin peroxidase from Bacillus megaterium and its application in bioremediation. CIBTech J. Microbiol, 3, 22-28.
Pita, M. P., Reviejo, A. J., De Villena, F. M., & Pingarrón, J. M. (1997). Amperometric selective biosensing of dimethyl-and diethyl dithiocarbamates based on inhibition processes in a medium of reversed micelles. Analyticachimicaacta, 340(1-3), 89-97. 00552-1
View ArticlePohanka, M., Vlček, V., Kuča, K., Bandouchová, H., & Pikula, J. (2010). Pesticide sorption in typical Central European soils evaluated using a photometric microplate assay based on acetylcholinesterase inhibition. Journal of Applied Biomedicine, 8, 41-46.
View ArticlePriest, F. G. (1977). Extracellular enzyme synthesis in the genus Bacillus. Bacteriological Reviews, 41(3), 711-711. PMid:334155
View Article PubMed/NCBIPriya, D., kumar, D. J. M., & Kalaichelvan, P. T. (2014). Optimization and production of Extracellular alkaline phosphatase from Bacillus megaterium. Internationl Journal of ChemTech Research, 6(9), 4251-4258.
Rao, P. R., & Kavya, P. R. (2014). Production, isolation and purification of peroxidase using Bacillus Subtilis. International Proceedings of Chemical, Biological and Environmental Engineering, 64, 21-27.
Raut, N., O'Connor, G., Pasini, P., & Daunert, S. (2012). Engineered cells as biosensing systems in biomedical analysis. Analytical and bioanalytical chemistry, 402(10), 3147-3159. PMid:22311427
View Article PubMed/NCBIRehena, F., Ventaksubbiah, L., & Naud, K. (1989). Preliminary studies on the production of thermostable amylase by mesophilic strain of Bacillus licheniformis. Chemical Microbiology Technology, 12, 8-13.
Rekha, K., Gouda, M. D., Thakur, M. S., & Karanth, N. G. (2000). Ascorbate oxidase based amperometric biosensor for organophosphorous pesticide monitoring. Biosensors and Bioelectronics, 15(9-10), 499-502. 00077-4
View ArticleSeki, A., Ortéga, F., & Marty, J. L. (1996). Enzyme sensor for the detection of herbicides inhibiting acetolactate synthase. Analytical Letters, 29(8), 1259-1271.
View ArticleSharma, D., Wangoo, N., & Sharma, R. K. (2021). Sensing platform for pico-molar level detection of ethyl parathion using Au-Ag nanoclusters based enzymatic strategy. Talanta, 221, 121267-121267. PMid:33076046
View Article PubMed/NCBIStark, J. R., Stewart, T. B., & Priest, F. G. (1982). Characterisation of extracellular α‐and β‐ amylases from Bacillus megaterium. FEMS Microbiology Letters, 15, 295-298. 90074-X
View ArticleTariq, A. L., Sudha, S., & Reyaz, A. L. (2016). Isolation and screening of bacillus species from sediments and application in bioremediation. Int.J.Curr.Microbiol.App.Sci.5(6, 916-924.
View ArticleVidal, J. C., Bonel, L., & Castillo, J. R. (2008). A Modulated Tyrosinase Enzyme‐Based Biosensor for Application to the Detection of Dichlorvos and Atrazine Pesticides. Electroanalysis: An International Journal Devoted to Fundamental and Practical Aspects of Electroanalysis, 20(8), 865-873.
View ArticleVishnu, T. S., Soniyamby, A. R., Praveesh, B. V., & Hema, T. A. (2014). Production and optimization of extracellular amylase from soil receiving kitchen waste isolate bacillus sp. VS 04. World Applied Sciences Journal, 29, 961-967.
Weibull, C., Beckman, H., & Bergström, L. (1959). Localization of enzymes in Bacillus megaterium, strain M. Microbiology, 20(3), 519-531. PMid:13664899
View Article PubMed/NCBIWood, D. A., & Tristram, H. (1970). Localization in the cell and extraction of alkaline phosphatase from Bacillus subtilis. Journal of Bacteriology, 104(3), 1045-1051. PMid:16559076
View Article PubMed/NCBIWu, L., Zhou, M., Liu, C., Chen, X., & Chen, Y. (2021). Double-enzymes-mediated Fe2+/Fe3+ conversion as magnetic relaxation switch for pesticide residues sensing. Journal of Hazardous Materials, 403, 123619-123619. PMid:32827859
View Article PubMed/NCBIWu, S., Zhang, L., Qi, L., Tao, S., Lan, X., Liu, Z., & Meng, C. (2011). Ultra-sensitive biosensor based on mesocellular silica foam for organophosphorous pesticide detection. Biosensors and Bioelectronics, 26(6), 2864-2869. PMid:21185711
View Article PubMed/NCBIXiao, Z., Storms, R., & Tsang, A. (2006). A quantitative starch? Iodine method for measuring alpha-amylase and glucoamylase activities. Analytical Biochemistry, 351(1), 146-148. PMid:16500607
View Article PubMed/NCBIYang, C., Lim, W., & Song, G. (2020). Mediation of oxidative stress toxicity induced by pyrethroid pesticides in fish. Comparative Biochemistry and Physiology Part C: Toxicology & Pharmacology, 234, 108758. PMid:32289527
View Article PubMed/NCBIYang, X., Dai, J., Zhao, S., Li, R., Goulette, T., Chen, X., & Xiao, H. (2018). Identification and characterization of a novel carboxylesterase from Phaseolus vulgaris for detection of organophosphate and carbamates pesticides. Journal of the Science of Food and Agriculture, 98(13), 5095-5104. PMid:29604085
View Article PubMed/NCBIYunhe, Q., Sun, Q., Xiao, F., Shi, G., & Jin, L. (2010). Layer-by-layer self-assembled acetylcholinesterase/PAMAM-Au on CNTs modified electrode for sensing pesticides. Bioelectrochemistry,77(2),139-144. PMid:19733130
View Article PubMed/NCBIZheng, J. Y., Wang, J., Zhou, S. S., Li, X. J., Ying, X. X., & Wang, Z. (2017). A stereoselective esterase from Bacillus megaterium: Purification, gene cloning, expression and catalytic properties. Protein Expression and Purification, 136, 66-72. PMid:26518366
View Article PubMed/NCBI