Feng-Jie Jin
Tel.: +86-25-8542-7210, Fax: +86-25-8542-7210,
Email: jinfj@njfu.edu.cn.
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 2
Page No: 645-651
Feng-Jie Jin
Tel.: +86-25-8542-7210, Fax: +86-25-8542-7210,
Email: jinfj@njfu.edu.cn.
Bao-Teng Wang, Zhi-Min Zhang, Miao Zhuang, Long Jin, Xing-Ye Yu, Yun-Jia Zhu, and Feng-Jie Jin*
Co-Innovation Center for Sustainable Forestry in Southern China, College of Biology and the Environment, Nanjing Forestry University, 159 Longpan Road, Nanjing 210037, China
Bart?omiej Zieniuk(bartlomiej_zieniuk@sggw.pl)
Yi Li(liyi396679338@gmail.com)
Subhash C Minocha(Subhash.Minocha@unh.edu)
Feng-Jie Jin, Effects of SclR on metabolite production in Aspergillus oryzae(2019) SDRP Journal of Food Science & Technology 4(2)
Aspergillus oryzae, an industrially important fungus utilized for the traditional fermentative manufacturing, produces diverse metabolites and enzymes during fermentation. In our previous study, we have ever identified and characterized a basic helix-loop-helix (bHLH) transcription factor, SclR, and found that the transcription factor is importantly involved in the conidiation, sclerotial formation, and hyphal morphology in A. oryzae. In this study, we further analyzed the effect of SclR on metabolites production in A. oryzae. The control strain and ∆sclR strain were cultivated in dextrin-polypeptone-yeast extract (DPY) liquid medium at 30 °C for 3 days and metabolites were extracted from strains. Liquid chromatography–mass spectrometry (LC-MS) analysis result showed that a total of 1,855 precursor molecules were obtained in the positive ion mode and 2,475 precursor molecules were obtained in the negative ion mode. Comparative analysis of the metabolome indicated that 259 were differentially expressed metabolites. Among of them, 161 metabolites were increased by 1.5-fold, and 98 metabolites were decreased by 1.5-fold in an sclR gene deletion strain compared to the control. Further identification of differentially expressed metabolites revealed that the production level of some organic acids, amino acids, and sugar alcohols were markedly changed in ∆sclR strain relative to control strain. Especially, some organic acids were identified to be intermediates in the citric acid cycle, which is a key metabolic pathway that connects carbohydrate, fat, and protein metabolism. The result also implied that the SclR is potentially involved in energy metabolism.
Keywords :Aspergillus oryzae · Transcription factor · SclR · Metabolite · Citric acid cycle
The filamentous fungus Aspergillus oryzae is an industrially important fermentative production strain. It has a strong capacity of producing and secreting protein and therefore A. oryzae has also been used for commercial enzyme production (Thammarongtham et al., 2018; Jujjavarapu and Dhagat 2019). Because of the industrial significance of A. oryzae, its genome has been completely sequenced (Machida et al. 2005; Kobayashi et al. 2007) and functional genes have also been continuously studied and explored.
The basic helix-loop-helix (bHLH) proteins belong to a superfamily of transcription factors which are often involved in the regulation of growth and differentiation. They have been well characterized and play an important role in the regulation of neurogenesis, cell proliferation and differentiation, myogenesis, sex determination, cell lineage determination, and other essential processes in organisms ranging from yeasts to mammals (Ledent and Vervoort 2001; Wu and Rastinejad 2017). In Aspergillus species, some bHLH transcription factors have been identified and characterized. For example, StuA, AnBH1, and DevR have been described in Aspergillus nidulans and they are importantly involved in the regulation of penicillin biosynthesis, sexual and asexual development, and regulation of development and cell cycle progression, respectively (Dutton et al. 1997; Caruso et al. 2002; Tüncher et al. 2004; Valiante et al. 2016). Among of them, the function of DevR was further characterized in A. oryzae,and found that DevR potentially interacted with promoter of amyR, which encodes a transcription factor involved in amylase production, and significantly affects polysaccharide metabolism in A. oryzae (Zhuang et al., 2019). In our previous study, in A. oryzae, we have ever identified and characterized the two novel bHLH transcription factors, SclR and EcdR, and they co-regulated the conidiation and sclerotial formation by forming competitively heterodimers (Jin et al. 2009, 2011a, 2011b). Overexpression of sclR led to abnormal hyphal morphology and it has been reported that sclR overexpression promoted the formation of heterokaryotic sclerotia in A. oryzae (Wada et al. 2014). In addition, loss of the sclR gene function led to rapid protein degradation and cell-lysis in liquid medium. Comparative intracellular proteomics indicated that SclR is importantly involved in carbohydrate metabolism, especially glycolysis pathway, in A. oryzae (Jin et al., 2018).
Metabolomics is a useful and important tool in various fields, such as agriculture, food science, and microbiology (Wishart 2008; Lee and Son 2013). Metabolite profiling aims to monitor all metabolites in a sample and metabolomic strategies have also been used to investigate metabolite changes caused by filamentous fungi during fermentation (Kim et al., 2010; Lee et al., 2016).
In this study, to further explore the functions of SclR, a comparative metabolomics analysis of the intracellular metabolites of a sclR deletion strain and a control strain was performed. The result indicated that SclR was involved in the citric acid cycle, which is the most important central pathway connecting almost all the individual metabolic pathways. Therefore, it was concluded that the SclR has an importantly role in the industrial production and utilization of A. oryzae.
Fungal Strains, Media, and Cultivation
The Aspergillus oryzae RIB40 is a wild type strain, the sclR gene deletion strain, ∆sclR (∆ku70::ptrA, ∆sclR::pyrG) (Jin et al. 2009) and the control RkuN16ptr1(pyrG) (∆ku70::ptrA, ∆pyrG,pyrG+) were used for the metabolites analysis. DPY medium (2% dextrin, 1% polypeptone, 0.5% yeast extract, 0.5% KH2PO4, and 0.05% MgSO4·7H2O, pH 5.8) was used for the liquid cultivation of A. oryzae. A total of 106 conidia of each strain were cultivated in 100 ml of DPY liquid medium at 30 °C for 3 days and two repeated experiments were performed for each strain. The mycelia of the two strains were collected by filtration through a Miracloth (Calbiochem, Darmstadt, Germany), washed and then rapidly frozen in liquid nitrogen for future use in the metabolites extraction procedure.
Sample preparation
60 mg mycelia of the strain were transferred into 2 mL centrifuge tubes, and then 500μL of methanol (pre-cooled at -20℃), 500μL deionized water (ddH2O) (4℃), and 100mg glass beads were added and mixed. Samples were further treated for 5 min by liquid nitrogen and thaw at room temperature, then put tubes in the high flux organization grinding apparatus (SCIENTZ-48, SCIENTZ, China), 70 Hz 2min. Samples were centrifuged and blow-dried by vacuum concentration, and then dissolved with 300ul methanol aqueous solution (1:1). Finally, the samples were filtered by 0.22 um membrane (Jin Teng, China) and ready for LC - MS detection.
LC-MS/MS analysis and data processing
Chromatographic separation was accomplished in an Acquity UPLC system equipped with an ACQUITY UPLC® HSS T3 (150×2.1 mm, 1.8 μm; Waters Corporation, Eschborn, Germany) column maintained at 40℃. The temperature of the autosampler was 4℃. Gradient elution of analytes was carried out with 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B) at a flow rate of 0.25 mL/min. Injection of 3μL of each sample was done after equilibration. An increasing linear gradient of solvent B (v/v) was used as follows: 0~1 min,2% B;1~9.5 min,2%~50% B;9.5~14min,50%~98% B;14~15 min,98% B;15~15.5 min,98%~2% B;15.5~17 min,2%. The ESI-MS experiments were executed on the Thermo LTQ-Orbitrap XL mass spectrometer (Thermo Scientific, San Jose, CA, USA) with the spray voltage of 4.8 kV and -4.5 kV in positive and negative modes, respectively. Sheath gas and auxiliary gas were set at 40 and 15 arbitrary units, respectively. The capillary temperature was 325 ℃. The voltages of capillary and tube were 35 V and 50 V, -15 V and -50 V in positive and negative modes, respectively. The Orbitrap analyzer scanned over a mass range of m/z 50-1000 for full scan at a mass resolution of 60 000. Data dependent acquisition (DDA) MS/MS experiments were performed with CID scan. The normalized collision energy was 30 eV. Dynamic exclusion was implemented with a repeat count of 2, and exclusion duration of 15 s.
Proteowizard software (v3.0.8789) was used to convert the obtained raw data into mzXML format (XCMS input file format) (Smith et al., 2006) and R (v3.3.2) XCMS package was used for peaks recognition. Finally, the data which includes mass to charge ratio (m/z), retention time and peak intensity, were obtained. The identification of metabolites was performed according to the database: Human Metabolome Database (HMDB) (http://www.hmdb.ca), Metlin (http://metlin.scripps.edu), Massbank (http://www.massbank.jp/), LipidMaps (http://www.lipidmaps.org), Mzclound (https://www.mzcloud.org). A KEGG pathway analysis of the identified differentially expressed metabolites was done.
Differentially expressed metabolites between ∆sclR and control strains
To further analyze the potential functions of the bHLH transcription factor SclR, an analysis and comparison of intracellular metabolites between sclR gene deletion strain and control strain were performed by using the liquid chromatography–mass spectrometry (LC-MS). In this study, two repeated culture experiments were performed for each strain. The control strain and ∆sclR strain were cultivated in dextrin-polypeptone-yeast extract (DPY) liquid medium at 30 °C for 3 days and metabolites were extracted from strains. The heatmap and hierarchical cluster analysis of the metabolites was performed (Fig. 1A). LC-MS analysis result showed that 1,855 precursor molecules were obtained in the positive ion mode and 2,475 precursor molecules were obtained in the negative ion mode. Comparative analysis of the metabolome indicated that a total of 259 metabolites displayed different production level. Among of them, 161 metabolites were increased by 1.5-fold, and 98 metabolites were decreased by 1.5-fold in an sclR-disrupted strain compared to the control (Fig. 1B-C; Table S1 in Supplementary material). A Benjamini-Hochberg corrected P-value < 0.05 was used as the threshold to denote the significance.
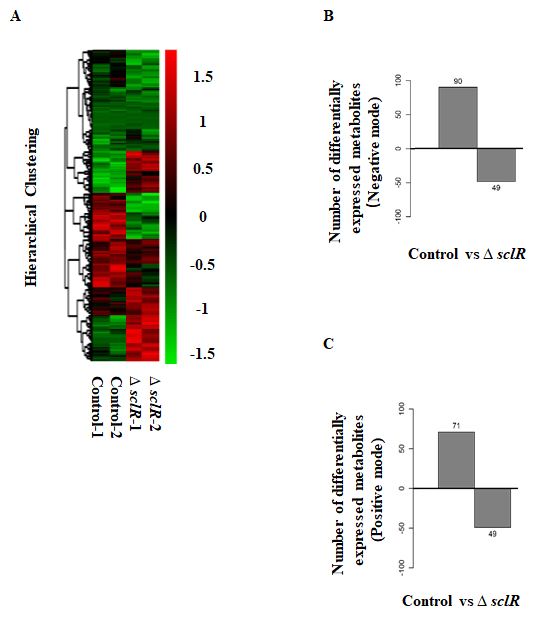
Fig. 1: LC-MS data analysis. (A) Hierarchical cluster analysis of the metabolites. Each strain was cultivated in DPY liquid medium at 30 °C for 3 days. The mycelia were collected for further LC-MS analysis. The hierarchical clustering diagram of the relative quantitative values of metabolites was showed. The relative quantitative values of metabolites were normalized and clustered, with red representing the highly expressed metabolites and blue representing the poorly expressed metabolites. Differentially expressed metabolites obtained in the negative (B) and positive (C) ion modes between ∆sclR and control strains. A total of 161 metabolites were increased, and 98 metabolites were decreased in expression level in an sclR gene deletion strain compared to the control (Fold_change ≥1.5 or ≤0.667; p-value < 0.05). Control, RkuN16ptr1(pyrG) strain; ∆sclR, sclR gene deletion strain.
Identification of partially differential expressed metabolites
The identification of metabolites was performed according to the accurate molecular weight, and then the MS/MS fragment was evaluated according to the database as mentioned in Materials and Methods section, as well as the self-built standards database to confirm and annotate the obtained metabolites. According to these results, we found that some metabolites were identified to be organic acids, amino acids, sugar alcohols, etc. (Fig. 2A-C). Among of them, the expression level of inositol was significantly increased, while other metabolites were obviously decreased in the sclR deletion strain compared to the control (Fig. 2). Interestingly, according to the KEGG pathway analysis, some organic acids, such as citric acid, ketoglutaric acid and fumaric acid, were found to be involved in the citric acid cycle, which is a key metabolic pathway that connects carbohydrate, fat, and protein metabolism (Akram 2014). Most of these metabolic processes are related to energy metabolism. Therefore, the citric acid cycle is the most important metabolic pathway for the energy supply to the growth of strain. Moreover, in our previous study, the expression level of citrate synthase (AO090009000568), which is one of the major enzymes regulated citric acid cycle and catalyzes the condensation of oxaloacetate with acetyl CoA to form citrate, was also found to be markedly decreased in the sclR gene deletion strain compared to the control by using isobaric tandem mass tag (TMT) labeling for quantification (Jin et al., 2018). In addition, it has been reported that an important link between intermediates of the citric acid cycle and the regulation of hypoxia-inducible factor. Hypoxia-inducible factor plays a role in the regulation of oxygen homeostasis, and is a transcription factor that targets glucose utilization, iron transport and apoptosis (Koivunen et al., 2007). In protein catabolism, proteins are broken down into amino acids and their carbon skeletons may enter the citric acid cycle as intermediates (e.g. alpha-ketoglutarate derived from glutamate or glutamine), having an effect on the cycle. At the same time, several of the citric acid cycle intermediates are used for the synthesis of important compounds, for example, the carbon skeletons of many amino acids are made from citric acid cycle intermediates (Akram 2014). Consistent with this, in these differential expression metabolites, we also identified two amino acids: L-tyrosine and L-leucine. The expression level of the two amino acids was also importantly decreased in the sclR deletion strain (Fig. 2B). Additionally, some sugar alcohols (Arabitol, mannitol, and inositol), which are organic compounds and typically derived from sugars, were also notably changed in production level in the sclR deletion strain relative to control strain (Fig. 2C). Except for these, the expression of nicotinic acid was also slightly decreased in the sclR deletion strain (data not shown). In our previous research, we found that the SclR is importantly involved in carbohydrate metabolism, especially glycolysis pathway, and even fatty acid degradation (Jin et al., 2018). According to our research, most of the enzymes involved in glycolysis and other carbohydrate metabolic pathways are down-regulated in the sclR-disrupted strain. In addition, the API-ZYM assay results also indicated that three enzymes (β-galactosidase, α-glucosidase, and α-mannosidase) were significantly less active in the ∆sclR strain than in the control (Jin et al., 2018). Glycolysis of glucose to pyruvate is also accompanied by energy production in the form of ATP and NADH, implying that energy production may also be restricted accordingly in the sclR-disrupted strain. In addition, the pyruvate produced by glycolysis is an important intermediate in the conversion of carbohydrates into citric acid cycle. These results are consistent with our current study, demonstrating that SclR is importantly involved in energy metabolism. This is also one of the possible reasons for the abnormal mycelial morphology of the sclR gene deletion strain. In this study, the LC-MS/MS experiment and comparative analysis of metabolites suggested that SclR is significantly involved in citric acid cycle and participated in energy metabolism, further revealing that the function of SclR in the growth and development stage of A. oryzae strain.
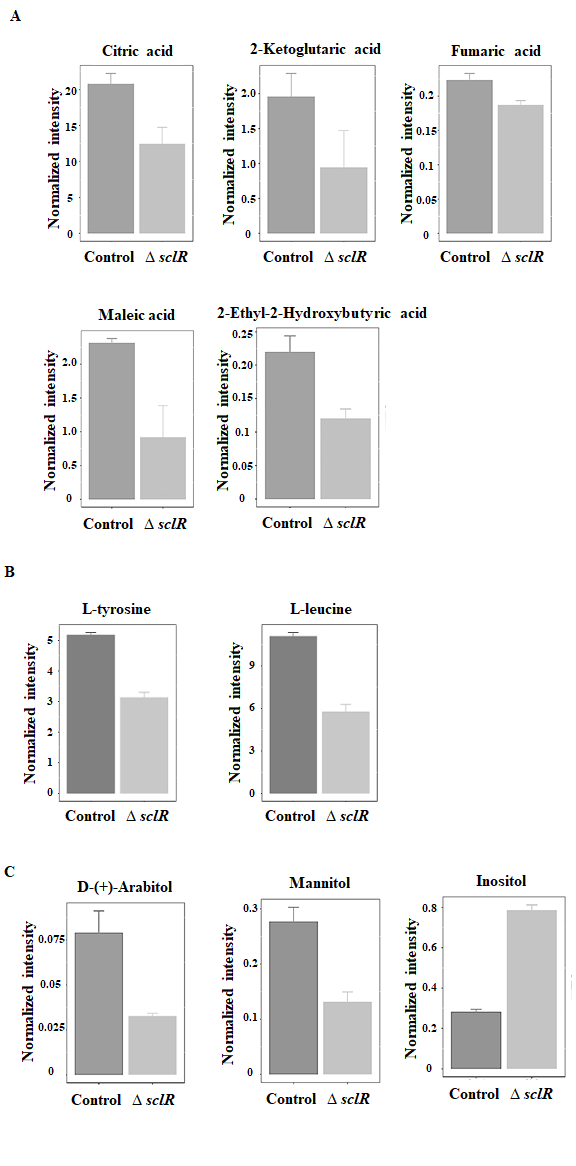
Fig. 2: Identification of some differentially expressed metabolites. Some differentially expressed metabolites were identified to be organic acids (A), amino acids (B), and sugar alcohols (C) (p-value < 0.05). Control, RkuN16ptr1(pyrG) strain; ∆sclR, sclR gene deletion strain.
Table S1-1 Differentially expressed metabolites obtained in the negative ion mode
(https://www.siftdesk.org/articles/images/476/Table-S1-1.pdf)
Table S1-2 Differentially expressed metabolites obtained in the positive ion mode
(https://www.siftdesk.org/articles/images/476/Table-S1-2.pdf)
This study was supported by the Natural Science Foundation of China (31570107), and the project was funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Akram M. 2014. Citric acid cycle and role of its intermediates in metabolism. Cell Biochem Biophys. 2014 Apr;68(3):475-8. PMid:24068518
View Article PubMed/NCBICaruso ML, Litzka O, Martic G, Lottspeich F, Brakhage AA (2002) Novel basic-region helix-loop-helix transcription factor (AnBH1) of Aspergillus nidulans counteracts the CCAAT-binding complex AnCF in the promoter of a penicillin biosynthesis gene. J Mol Biol 323:425–439. 00965-8
View ArticleDutton JR, Johns S, Miller BL (1997) StuAp is a sequence-specific transcription factor that regulates developmental complexity in Aspergillus nidulans. EMBO J 16:5710–21. PMid:9312029
View Article PubMed/NCBIJin FJ, Han P, Zhuang M, Zhang ZM, Jin L, Koyama Y. 2018. Comparative proteomic analysis: SclR is importantly involved in carbohydrate metabolism in Aspergillus oryzae. Appl Microbiol Biotechnol 102(1):319-332. PMid:29098410
View Article PubMed/NCBIJin FJ, Nishida M, Hara S, Koyama Y. 2011b. Identification and characterization of a putative basic helix-loop-helix transcription factor involved in the early stage of conidiophore development in Aspergillus oryzae. Fungal Genet Biol 48: 1108–15. PMid:22008745
View Article PubMed/NCBIJin FJ, Takahashi T, Matsushima K, Hara S, Maruyama J, Kitamoto K, Koyama Y (2011a) SclR, a basic helix-loop-helix transcription factor, regulates hyphal morphology and promotes sclerotial formation in Aspergillus oryzae. Eukaryot Cell 10:945–955. PMid:21551246
View Article PubMed/NCBIJin FJ, Takahashi T, Machida M, Koyama Y. 2009. Identification of bHLH-type transcription regulator gene by systematically deleting large chromosomal segments in Aspergillus oryzae. Appl Environ Microbiol 75:5943–5951. PMid:19633118
View Article PubMed/NCBIJujjavarapu SE, Dhagat S. 2019. Evolutionary Trends in Industrial Production of α-amylase. Recent Pat Biotechnol. 13(1):4-18. PMid:30810102
View Article PubMed/NCBIKim AJ, Choi JN, Kim J, Park SB, Yeo SH, Choi JH, Lee CH. 2010. GC-MS based metabolite profiling of rice koji fermentation by various fungi. Biosci. Biotechnol. Biochem. 74:2267–2272. PMid:21071848
View Article PubMed/NCBIKobayashi T, Abe K, Asai K, Gomi K, Juvvadi PR, Kato M, Kitamoto M, Takeuchi M, Machida M (2007) Genomics of Aspergillus oryzae. Biosci Biotechnol Biochem 71:646–670. PMid:17341818
View Article PubMed/NCBIKoivunen P, Hirsilä M, Remes AM, Hassinen IE, Kivirikko KI, Myllyharju J. 2007. Inhibition of hypoxia-inducible factor (HIF) hydroxylases by citric acid cycle intermediates: possible links between cell metabolism and stabilization of HIF. J. Biol. Chem. 282 (7): 4524–32.
View ArticleLedent V, Vervoort M. 2001. The basic helix-loop-helix protein family: Comparative genomics and phylogenetic analysis. Genome Res 11:754–770. PMid:11337472
View Article PubMed/NCBILee da E, Lee S, Jang ES, Shin HW, Moon BS, Lee CH. 2016. Metabolomic Profiles of Aspergillus oryzae and Bacillus amyloliquefaciens During Rice Koji Fermentation. Molecules. 14:21(6). pii: E773. doi: 10.3390/molecules21060773.
View ArticleLee MY. and Son GH. 2013. Liquid chromatography-mass spectrometry-based chemotaxonomic classification of Aspergillus spp. and evaluation of the biological activity of its unique metabolite, neosartorin. J. Microbiol. Biotechnol. 23:932–941. PMid:23711526
View Article PubMed/NCBIMachida M, Asai K, SanoM, Tanaka K, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, Abe K, Gomi K, Horiuchi H, Kitamoto K, Kobayashi T, Takeuchi M, Denning DW, Galagan JE, Nierman WC, Yu J, Archer DB, Bennett JW, Bhatnagar D, Cleveland TE, Fedorova ND, Gotoh O, Horikawa H, Hosoyama A, Ichinomiya M, Igarashi R, Iwashita K, Juvvadi PR, Kato M, Kato Y, Kin T, Kokubun A, Maeda H, Maeyama N, Maruyama J, Nagasaki H, Nakajima T, Oda K, Okada K, Paulsen I, Sakamoto K, Sawano T, Takahashi M, Takase K, Terabayashi Y, Wortman JR, Yamada O, Yamagata Y, Anazawa H, Hata Y, Koide Y, Komori T, Koyama Y, Minetoki T, Suharnan S, Tanaka A, Isono K, Kuhara S, Ogasawara N, Kikuchi H. 2005. Genome sequencing and analysis of Aspergillus oryzae. Nature 438:1157–1161. PMid:16372010
View Article PubMed/NCBISmith CA, Want EJ, O'Maille G, Abagyan R, and Siuzdak G. 2006. XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Analytical chemistry. 78, 779-787.
View ArticleThammarongtham C, Nookaew I, Vorapreeda T, Srisuk T, Land ML, Jeennor S, Laoteng K. 2018. Genome Characterization of Oleaginous Aspergillus oryzae BCC7051: A Potential Fungal-Based Platform for Lipid Production. Curr Microbiol. 2018 Jan;75(1):57-70.
View ArticleTüncher A, Reinke H, Martic G, Caruso ML, Brakhage AA. 2004. A basic-region helix-loop-helix protein-encoding gene (devR) involved in the development of Aspergillus nidulans. Mol Microbiol 52(1):227-41. PMid:15049823
View Article PubMed/NCBIWada R, Jin FJ, Koyama Y, Maruyama J, Kitamoto K (2014) Efficient formation of heterokaryotic sclerotia in the filamentous fungus Aspergillus oryzae. Appl Microbiol Biotechnol 98:325–334. PMid:24201891
View Article PubMed/NCBIWishart DS. 2008. Metabolomics: Applications to food science and nutrition research. Trends Food Sci. Technol. 19: 482–493.
View ArticleWu D, Rastinejad F. 2017. Structural characterization of mammalian bHLH-PAS transcription factors. Curr Opin Struct Biol. 43:1-9. PMid:27721191
View Article PubMed/NCBIValiante V, Baldin C, Hortschansky P, Jain R, Thywißen A, Straßburger M, Shelest E, Heinekamp T, Brakhage AA. 2016. The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. Mol Microbiol 102(2):321-335. PMid:27393422
View Article PubMed/NCBIZhuang M, Zhang ZM, Jin L, Wang BT, Koyama Y, and Jin FJ. 2019. The bHLH transcription factor DevR significantly affects polysaccharide metabolism in Aspergillus oryzae. Appl Environ Microbiol. doi: 10.1128/AEM.00089-19. Accepted.
View Article