Zhuang Weijian
Email: weijianz@fafu.edu.cn
© 2019 Sift Desk Journals. All Rights Reserved
VOLUME: 4 ISSUE: 9
Page No: 986-1005
Zhuang Weijian
Email: weijianz@fafu.edu.cn
Shahid Ali Khan 1,2 and Zhuang Weijian 1,2,3*
1Fujian Provincial Key Laboratory of Plant Molecular and Cell Biology, State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops
2College of Crop Science Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China
3College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, Fujian 350002, China
Amanda Juan Chen(amanda_j_chen@163.com)
Ye-Lin Shao(shzoyelin@126.com)
Luca Morelli(luca.morelli@unipi.it)
Shahid Ali Khan, Zhuang Weijian, Obliging Tactics to Mitigate the Intricate Problem of Aflatoxin Contamination in Peanut: A Review(2019)Journal of Food Science & Technology 4(9)p:986-1005
Peanut (Arachis hypogaea L.) grown throughout the globe for its protein and oil contents. Its kernels are consumed as raw, boiled or roasted, and also in the form of culinary oil. Being a rich source of human diet (antioxidants, minerals and vitamins), animal feed (oil pressings, green straw and pods), industrial raw material (oil cakes and fertilizer), and soil fertility (atmospheric nitrogen fixation), peanut is a brilliant cash crop for both domestic markets as well as international trade. Having crystal clear importance in food and feed security peanut products are severely contaminated by aflatoxins (AFs). AFs produced mainly by Aspergillus flavus (A. flavus) and Aspergillus parasiticus (A. parasiticus), are secondary metabolites that jeopardize both human and animal health. There is no magic bullet found yet to solve this problem. Several techniques have been tested to minimize and control AFs contamination including different physical, chemical, and biological preventions. Many biological control agents, including nontoxigenic fungal strains, yeasts, and bacteria have been applied and considerable achievements gained. However, for complete eradication, a surge of studies is required to deeply investigate this intricate problem at gene and nucleotide levels and discover a permanent solution through elucidating its mechanism. The current review is focused on knowledge about A. flavus, its optimal growth conditions, growth promoting factors, factors affecting the level of AFs production, AFs biosynthesis pathway, mechanisms involved in resistance against fungal infection, various techniques and some simple precautionary recommendations to minimize AFs production.
Fig. Graphical abstract
Key words: Aflatoxins, Aspergillus flavus, Aspergillus parasiticus, Peanut, Secondary metabolites
Peanut (Arachis hypogaea) is grown in most parts of the world and more than 100 countries share its cultivation. It is cultivated mostly in the arid and semi-arid regions of the globe, where the two major growing partners are China and India. The top ten producers of peanut globally are China, India, Nigeria, United States, Sudan, Myanmar, Tanzania, Argentina, Indonesia, and Senegal (Figure 1) [1, 2]. Peanut is a good source of protein and oil. The importance and value of peanut oil is extremely vibrant due to the presence of low level of saturated fat and rich in antioxidants, resveratrol, which are the main contributor in cardiovascular health. Peanut share more than $35 billion in world economy in terms of production. The integral part of global peanut cultivation (~ 95 %) belong to Asia and Africa, where farmers are doing cultivation with very negligible farming resources and the most part is under rain-fed conditions. This indicates that there is a great space for its yield and quality improvement in the future. Peanut kernels contain about 48 to 50 % oil and 25 to 28 % protein, providing a rich source of energy for a large group of the human community throughout the world. Beside a promising supplier of human food, peanut also provide significant share in animal feed in the form of haulms [3]. Peanut kernels are a good source of essential nutrients including various antioxidants, minerals, vitamins and more importantly a valuable source of mono-unsaturated fatty acids. It contains p-coumaric acid and resveratrol antioxidants, the important vitamin E and other B-complex groups of pantothenic acid, thiamine, vitamin B-6, niacin and folates, a decent source of poly phenols and flavonoids. Peanut being an important source of vital nutrients, recommend as a hope of ray to eradicate the micronutrient malnutrition in different part of the developing world. A best example is its role in Niger hunger, where peanut helped in saving thousands of lives.
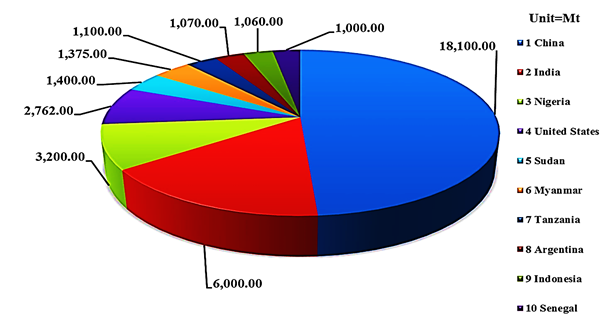
Fig. 1 Top ten peanut growers globally
1.2 Hot spot potential threat in peanut food products and feed consumption
No doubt peanut having a sheet anchor role in the world economy, but its production and quality are greatly threatened by aflatoxin (AFs) contamination. AFs, produced mainly by Aspergillus flavus (A. flavus) and Aspergillus parasiticus (A. parasiticus) as a secondary metabolite, are carcinogenic for both humans as well as animals [4, 5]. AFs contamination of crops is a worldwide food safety concern, refers to a group of four mycotoxins (B1, B2, G1 and G2). Chemical structures of these four types of AFs are somewhat related to each other (Figure 2). Strains of A. flavus show great variation in their ability to produce AFs. Toxigenic strains of A. flavus typically produce only two types of AFs, B1 and B2, but most strains of A. parasiticus can produce all the four toxins [6]. Since AFs are potential carcinogens, their quantity in food and feed is closely monitored and regulated in many countries. European Union has set a maximum level of 2µg kg-1 for B1 and 4µg kg-1 for total AFs in crops [7].
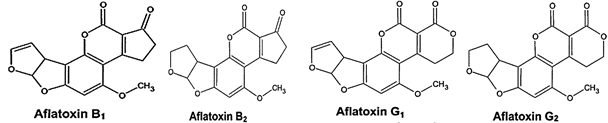
Fig. 2 Chemical structures of B1, B2, G1, and G2 AFs
Peanut production and quality are severely affected by A. flavus during both pre- and post-harvest conditions [8, 9]. Prior to harvesting A. flavus infection frequently occurs when the pods come in direct connection with its spores in the soil. The population density of the fungus, moisture level and temperature of soil at different levels of pods development are the various factors responsible for the intensity of A. flavus infection [10]. It is very difficult and almost impossible to remove the AFs once the food item is contaminated with it, even with different types of food processing and cooking practices. Roasting, however, is found helpful up to some extent to minimize the AFs contamination in peanut. But, it is crystal clear that the prevention of AFs contamination is better than the application of various costly strategies which are applied to remove the contamination. The most promising footstep to overcome this problem is growing resistant cultivars, but in case of peanut development of resistant cultivars to AFs contamination is a challenging job due to its very young genomic research. Currently improvement in peanut is mainly dependent on a few promising genetic cultivars, different cultural management strategies and various techniques to control and minimize the intensity of disease and pest infestation. Several cultivars of peanut were developed through conventional breeding systems but their resistance level is still not enough to completely prevent AFs contamination. Now a day the contamination of various agro-products by AFs is the hot spot threat in most of the developing countries to both human as well as animal, because of having worse effect on food and feed safety and security [11, 12]. It is also the main economic anxiety in world peanut industry, because due to the concern of AFs contamination extra regulatory principles are needed to apply, which further raise the cost of various peanut products. To minimize and eliminate the presence of AFs, there is a great need to deeply understand the interactions between the peanut and Aspergillus at different growth stages under various environmental conditions. Resistant varieties being most important source to overcome and minimize AFs contamination in peanut, still available cultivars are negligible. Till now the knowledge about the mechanism which cause resistance to Aspergillus at molecular level is very young [12]. So, an alternative way is to apply a combination of cultural, biological and chemical methods to minimize AFs production in peanut.
Peanut remained most preferable host crop for A. flavus infection and AFs contamination effecting its kernel very severely (Figure 3). The most suitable and optimal growth conditions for AFs production are in the regions where the temperature remains hot and humid [13, 14]. Beside optimal growth temperature, the production level of AFs is also enhanced by various factors like damage or stress in pre-harvest condition, various activities of insects, type of soil and lack of proper storage conditions [15]. AFs, especially B1 is responsible for hepatotoxicity and hepato-carcinogenicity. In the past the presence of AFs has been conformed in a number of agricultural products [16]. In case of peanut, A. flavus also have worse effect on the plant and is responsible for seed rots and molding of seeds, damping off at both pre- and post-emergence stages, affecting the viability level of seed and greatly reduces the strength of seedlings. No doubt the contamination of various food and feed products with AFs is a hot spot threat faced by food and feed safety and security throughout the globe, but this issue is comparatively more serious in developing countries [16, 17].
1.2.1 Worse experiences from the past about AFs contamination
The problem of food items contamination with AFs is not a new one to be solved, its prevention strategies has been started since 1960s. The main focus for its prevention was due to the famous “turkey X disease” which caused more than 100,000 deaths of turkey poults near London, England. The main cause of these deaths was AFs contaminated feed [18]. The focus on prevention strategies was further intensified by the 1970s outbreak in maize occurred in US, followed by a more serious outbreak in Kenya involved 317 cases caused 200 human deaths in 2004. These deaths were conformed due to direct aflatoxicosis caused by consumption of maize contaminated with AFs [16, 19, 20]. The consumption of contaminated food in low doses are the root cause of cancer and suppression of various immunological responses. Mostly the main and primary target of carcinogenic and toxic AFs remained the liver, where the end story includes the deadly liver cancer (Figure 4) [21]. AFs also have the ability to suppress the activity of those cells which are responsible for boosting human and animal immunity [22]. The level that how much these AFs are carcinogenic depends upon the amount and exposure time of the victim to it. Mainly due to these two factors two types of carcinogenic effects are found in the affected personals. (A) Acute illness and death, which is the result of consuming contaminated food containing very high levels of AFs. People died as a result of jaundice and liver failure, example is that of 2004 in Kenya where more than 200 people died. No animal species found yet to have resistance to acute toxic effects of AFs [16]. (B) Chronic illnesses or Cancers, caused due to the exposure to AFs of low level for a long time. International Cancer Research Institute (ICRI) categorizes AFs as a Class 1 carcinogen. Beside, AFs are also responsible for the interference in normal functioning of different cells, being capable of binding to various proteins, RNA and DNA to restrict their normal expression and thus causing cancer, mutations and necrosis in both human and animals [16]. AFs contamination is directly responsible for the economic loss in different crops especially in peanut, causing great reduction in market value. Increase occurs in the cost of product due to increase in application of healthcare and high regulatory principles. There are different regulatory principles applied by U.S. Food and Drug Administration (FDA) on levels of AFs. These levels are 20 ppb in food and feed items while 0.5 ppb in milk. These regulatory guidelines have put a great economic load of over US$932 million on agriculture worldwide due to crop losses caused by mycotoxigenic fungi including A. flavus. In extreme cases the food and feed items contaminated with AFs are completely rejected from the market. For example Africa alone pay more than US$670 million per annum to fulfill the EU principles for all food exports [12]. So, learning from the past worse experiences of AFs contamination, it is the order of the day to find out an environmental friendly solution to this distressing problem as soon as possible. The current review is focused on knowledge about A. flavus, its optimal growth conditions, growth promoting factors, AFs biosynthesis pathway, various techniques and some simple precautionary recommendations to minimize AFs production in peanut.
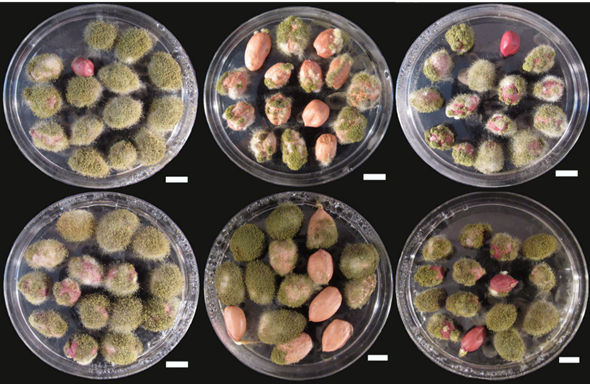
Fig. 3 Peanut kernels infected by A. flavus
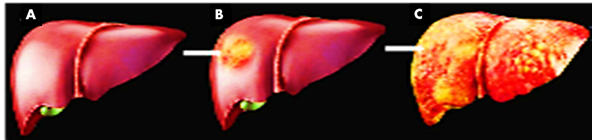
Fig. 4 A=human liver in normal shape, B= infected liver by AFs in initial stage, C= completely worn-out liver after AFs infections
2. Shape, ecology and geographical distribution of A. flavus
Due to indistinct differences in morphological and biochemical characteristics of Aspergillus species, its exact identification is a tedious job. Anyhow, A. flavus is believed to have velvety, brown or yellow to green mold with conidiophores of various lengths and mostly are pitted, rough and spiny either uniseriate or biseriate (Figure 5), covering the entire vesicle with pointed out phialides in every direction. Conidia are subglobose to globose, clearly echinulate, of having diameter within the range of 3.5 to 4.5 mm. Based on the characteristics of the produced sclerotia, A. flavus isolates can be distributed into two phenotypic types S and L. The S strain yields numerous small sclerotia having an average diameter less than 400 µm, while the L strain produces fewer but larger sclerotia. Within the S strain, some isolates, termed SB, produce only B AFs, whilst others, named SBG, produce both B and G AFs [23]. The S strain isolates have been referred to as a typical [24], producing microsclerotium [25] and A. flavus var. parvisclerotigenu [26]. The microsclerotial strains differs from A. flavus and therefore it has been suggested that they represent a taxon separated from A. flavus [26, 27]. Molecular phylogenetic analysis suggests that SB isolates are closely related to the A. flavus type culture and other L strain isolates [28].
A. flavus is distributed throughout the globe just like other related species of the Aspergillus genus. Its distribution is encouraged by airborne conidia as well as by insect activities. Similarly, humidity and other atmospheric elements also provide good support for mold vigorous growth. Water activity (aw) range of 0.86 to 0.96 also offers optimal growth condition to A. flavus. Normally A. flavus grow well at 37◦C but its growth can also be experienced with in the temperature range of 12 to 48◦C. The main strength of A. flavus due to which it wins the competition for substrate in plant/soil over other pathogens, is its ability to withstand a broad range of harsh environmental conditions. A. flavus form structures like mycelium or sclerotia, making it capable of overwintering. Then under favorable conditions, sclerotia either form further hyphae or asexual spores, called conidia, helping in the dispersion of fungus in the soil and air [29, 30].
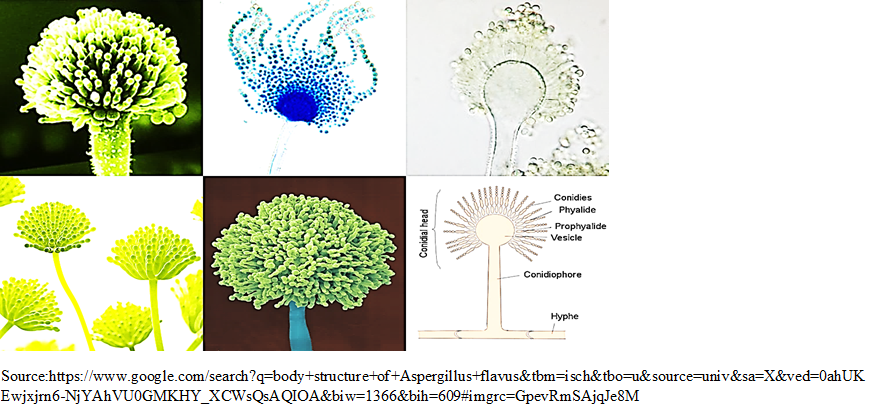
Fig. 5 Filamentous body structure of A. flavus
2.1 Factors affecting the level of AFs production
Fungal growth and AFs contamination are the consequences of interactions among the fungus, host and environment (Figure 6). Although some of the molecular mechanisms remains unclear, many biotic, abiotic, nutritional and environmental factors can affect the production of AFs [14]. Nutritional elements such as carbon source, nitrogen source, amino acids, fats, and trace elements can affect the production of AFs. Monosaccharide like Glucose, sucrose and maltose can promote the formation of AFs, though peptone, sorbitol and lactose cannot. However, the mechanism of the carbon source involved in the regulation of AFs biosynthetic pathway gene expression is still poorly understood. Nitrogen source affects the synthesis of AFs in different ways, when A. flavus lives in the medium of nitrate and nitrite, the levels of toxin varies [31]. Certain amino acids can also be counterproductive to the production of AFs. It has been found that tryptophan can inhibit AFs production, while tyrosine can promote the production of toxins [32]. It is reported that metal ions can affect both the growth of A. flavus and production of AFs at the cellular and molecular level [33]. Lipids make a great impact on the formation of AFs, it is not only a source of nutrition, but also a metabolic substrate [34] and signaling molecule [35] of the Fatty acyl-CoA. Some environmental factors such as temperature, pH, drought and other stresses are also believed to affect the production of AFs [36, 37]. Studies have shown that G-protein signaling catenation mediated by protein kinase A, can lead to AFLR gene transcription. This signaling pathway may respond to impact of the environment, thus affecting AFs biosynthesis [38]. When the temperature is close to 30◦C, AFs are most prone to be produced. The production of AFs is closely related to changes in pH, when the media is acidic, AFs can be formed, but in alkaline media the formation would be inhibited like the fungal growth, along with the secondary metabolites, such as sporulation and sclerotia formation [39]. Secondary metabolism and sporulation require similar environmental conditions. In addition, it is reported that the secondary metabolites are formed at the same time of sporulation [38, 40]. Mutant strains with no sporulation cannot produce AFs. Some compounds which can inhibit A. flavus producing spores are also been shown to inhibit the production of AFs. The oxidative stress can induce the production of AFs. After treated by tert-Butylhydroquinone, the production of AFs increase significantly [37]. There are also endogenous phytochemical constituents, capable of inhibiting AFs production of A. flavus, and the bioactivity resided in a complex of hydrolyzable tannins. These tannins can be hydrolyzed by a fungal tannase present in A. flavus, yielding gallic acid and ellagic acid, testing of which showed that gallic acid had potent inhibitory activity towards AFs biosynthesis [41].
The main contributors in increased level of AFs contamination are highly depended upon biotic (biological) and abiotc (environmental) factores providing optimal conditions for Aspergillus to produce AFs in high amount. The production of AFs is greatly incouraged by flood, heavy rain and poor storage conditions. The AFs production level is further increased by mechanical demage through various pest and different types of stress conditions. Variation in seasons, geographical conditions for example the penetration of fungal spores to different crop parts occures due to extreme variation in weather conditions, the kind of fungal strain present in that area, disturbance from other pest and organisams, moisture level of the soil and temperature are the elements which are involed in boosting AFs contaminaion [12, 40, 42, 43].
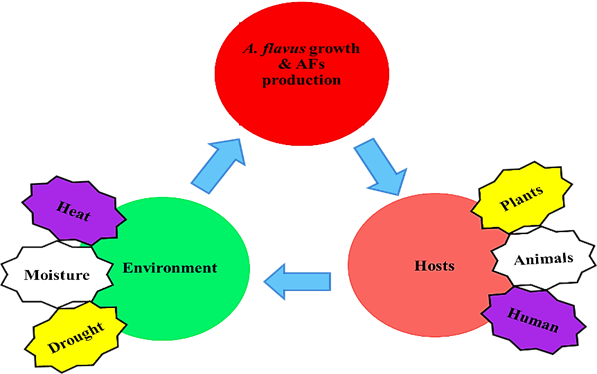
Fig. 6 Environment, A. flavus and Host interactions triangle
2.2 Schematic representation of AFs synthesis pathway
From the initial days of AFs identification, efforts have been started to control this problem [44-48]. The front-line discovery of a color mutant that stores the brick-red pigment, norsolorinic acid (NOR) in A. parasiticus, clear a milestone to understand the chemistry of AFs biosynthesis [49-52]. Since NOR discovery, the first one and stable AFs precursor in the AFs biosynthetic pathway [53-55], provided a vital role in identification of other intermediates in AFs synthesis pathway. It opened the opportunity for isolating the first AFs pathway gene which encoding a reductase for the conversion of NOR to its final product in the form of AFs [54, 56, 57]. After some important genes being cloned, the AFs pathway gene cluster was identified in A. flavus and A. parasiticus [58]. The discovery of the cluster stimulated renewed interest to understand AFs biosynthesis throughout the globe. Substantial progress has been gained in elucidating the biosynthetic pathway of AFs, the pathway intermediates, genes, their corresponding enzymes, and different regulatory mechanisms [59-72]. There are about 30 genes which putatively involved in AFs biosynthesis. Different studies have found that AFs synthesis pathway genes are clustered within a 75-kb region in A. flavus and A. parasiticus on chromosome III approximately 80 kb away from telomere [55, 72-78]. Throughout the globe, AFs synthesis pathway has been extensively studied by different scientists and they have got promising achievements, but the complete and sound basis of the AFs synthesis pathway are still ambiguous. A better way to deeply get into insights of the mechanism of AFs production, a comprehensive investigative approach must be applied, including classical gene cloning combined with modern whole genome sequencing approaches. AFs synthesis pathway is long and complex process governed by various other regulatory mechanism, but here it is expressed in a simple and short form, which can be easily understood (Figure 7). A chains of highly intricate oxidation-reduction reactions cause the formation of AFs. The given schematic diagram is currently the most putative scheme for AFs biosynthesis, which involves the formation of NOR from polyketide acting as a first and basic step towards AFs synthesis. This step is followed by the conversion of NOR to averantin (AVN) leading the pathway to its final products in the form of AFs production.
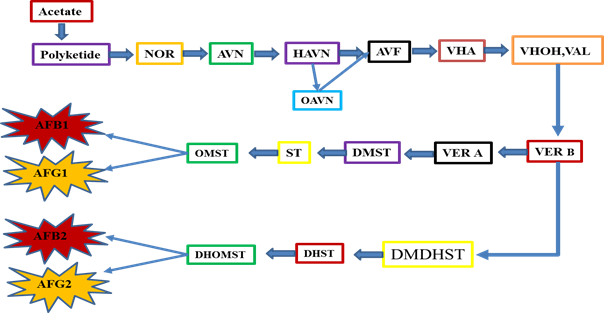
Fig.7 The schematic AFs synthetic pathway. Where NOR=Norsolorinic acid, AVN = Averantin, HAVN=5'-hydroxyaverantin, OAVN=oxoaverantin, AVF=averufin, VHA=versiconal hemiacetal acetate, VHOH(VAL)=versiconal, VER-B=versicolorin-B, VER-A=Aversicolorin-A, DMDHST=demethyldihydrosterigmatocystin, DMST=demethylsterigmatocystin, DHST=dihydrosterigmatocystin, ST=sterigmatocystin, OMST=O-methylsterigmatocystin, DHOMST=dihydro-o-methylsterigmatocystin, AFB1=aflatoxin B1, AFB2=aflatoxin B2, AFG1=aflatoxin G1, AFG2=aflatoxin G2
3. Genetics basis of resistance mechanisms
Resistance against A. flavus infection and its subsequent AFs production mechanisms are quantitative in nature [79]. In peanut, resistance to AFs may be attributed to three different levels: it may be due to resistance against fungal infection at pod wall, or resistance to seed invasion and colonization at seed coat, or it may be the result of resistance to AFs production in cotyledons of the seed. To infect peanut, A. flavus have to penetrate the pod wall and then pass through seed coat to get entree in to the cotyledons from where it derives its food and cause AFs contamination. Pod-shell structure and seed coat thickness, density of palisade cell layers, and presence of wax layers are the key traits contributing in resistance to pod infection and seed invasion and colonization [80]. Resistance to fungal infection can be achieved at three different levels. 1): In case of peanut, mostly AFs contamination occurs at pre-harvest stage and management practices of the crop. In this case resistant cultivars to fungal infection can play vital role in elimination of AFs contamination [81]. Resistance cultivars will provide great assistance in screening for resistance germplasm through using genomics-assisted breeding (GAB). 2): Seed coat thickness and its permeability contribute significantly in resistance against A. flavus infection acting outermost layer of seed defense [82]. Similarly, more compact arrangement of palisade like layer of testa accompanied by thicker waxy surface subsidizes resistance to A. flavus infection. Higher wax and cutin deposition are key elements contributing significantly in resistance level against A. flavus infection and AFs contamination, because wax content was found in significantly higher amount in resistant genotypes as compared to susceptible ones [83]. 3): Plants are endued with several inducible defense responses like lignification and cell wall cross-linking, hypersensitive response, phytoalexins, and production of active oxygen and numerous pathogenesis related proteins in response to pathogen attack [84]. Peanut seeds contain resveratrol, which is an antifungal secondary metabolite. It was noticed that the level of resveratrol was higher up to several folds even after three days of inoculation as compared with that of susceptible genotypes [85]. Keeping in view the above three factors during developing peanut commercial cultivars can greatly support the mission of AFs contamination elimination at both pre- and post-harvest stages of the crop.
4. Obliging techniques to reduce AFs contamination in peanut
Several techniques have been tested to minimize, prevent, eliminate or decontaminate different products from AFs contamination in peanut and other crops at various growth stages. Among these prevention measures the most affective one is to minimize and prevent AFs contamination at pre-harvest stage of the crop [86]. The main focus now is to minimize and prevent the contamination of AFs through fully exploring and understanding the interlationship between the crop and the fungus. Aspergillus infection is divided into three parts i.e., in the first part it damage the cell wall of the host through different enzymes, secondly, fungal mechinary is developed inside the host and at third stage the production of AFs occurs [87, 88]. Techniques which can minimize AFs contamination to a significant level includes using several biological agents, advanced cultural practices and more importantly the use of modern plant breeding techniques to develop resistant cultivars against A. flavus and other fungi responsible for AFs production.
4.1 Handy traits against AFs contamination
There are some traits the evaluation of which can control the AFs production to a significant level. In case of peanuts, A. flavus to get access to the cotyledons, from where it derive its nutritions, have to penetrate the pod wall and the seed coat [80]. Here pod-shell structure can play key role in resistance to pod infection, while resistance to kernel infection and colonization is generally physical, and mostly related with thickness, density level of palisade cell layers, absence of cavities and fissures, and due to wax layers. So the structure and characteristics of the pod can be used as a source of screening for AFs resistance traits. Throughout the world, drought stress before harvest is the main reason for AFs contamination in peanut. Drought resistance traits are promising as indirect selection tools for improving resistance to preharvest AFs contamination. Traits related to drought resistance were associated well with those related to preharvest AFs contamination under drought conditions. Besides, specific leaf area, relative water content, chlorophyll density and drought stress ratings are also the best traits can be used as indirect selection tools for lower preharvest AFs contamination. Breeding for drought tolerance using these traits as selection criteria may help to accelerate progress in developing resistance to preharvest AFs contamination [89]. Because of different evaluation criteria, selecting the resistance source directly is more complexed, usually resistance to AFs producing fungi can be divided in to three types i.e., bringing resistance to pod infection (pod wall), to kernel infection (seed coat), and resistance to AFs production (cotyledons) [90]. There are contradictory reports on the relationship among invitro seed colonization by A. flavus (IVSCAF)-resistance and under natural conditions in open field. Sources of all the three types of resistance have been reported, but mostly the results obtanied under IVSCAF were not conformed when tested under natural environemt [91]. This contrast in the resistance level evaluated under laboratory conditions compared with that of open field under natural environment make it a more complex, because the value of a resistant source mainly depends upon the stability of its resistance under various envirnmental conditions. The resistance level to pod infection has been found to be highly variable and the resistance found through IVSCAF-resistance is not the absolute one, because even the best sources show about 15 % seed colonization. Though, a few lines shown stable resistance but the resistance levels found are not very high [91], due to highly significant interections of genotype by environment for this trait.
4.2 Control of AFs contamination in susceptible crops
No doubt most promising elimination strategy of AFs is to develop resistant cultivars against it, but in case of susceptible one, we can also control and minimize its contamination following some simple precautionary procedures. The use of various fungicides which minimizes the fungal attack during growth season of the crop, providing proper storage conditions, including the use of different anti-mold preservatives, following reliable transport and distribution practices. A strict observation should be kept on aeration, temperature and moisture level of the storage facility, because these are the root cause of fungal growth. The entry of various insect under storage condition should be stopped. Proper storage conditions play key role in minimizing the risk posed by AFs contamination, because most of the products reach to consumers through a specific kind of storage condition. Following these simple precautionary measures, we can control and minimize the level of AFs production up to a significant level.
4.3 Control of AFs production via biological agents
Among the different techniques used for the control of AFs contamination, one and the most environment friendly is biological control or bio-control, which in simple words means control of life by life. Different bacterial species have been used to control and minimize the invasion of A. flavus and other related species which are responsible for AFs production. Among these bacterial species the famous one are Bacillus subtilis, Pseudomonas spp., Lactobacillis spp., Burkholderia spp. and Ralstonia spp., which inhibit Aspergillus and consequently AFs production under laboratory conditions [92]. Similarly, many Bacillus subtilis and Pseudomonas solanacearum strains are helpful to minimize AFs contamination when these were isolated from the maize grown soil other than the rhizosphere [93]. However, at field level in minimization of AFs contamination these bacterial strains were comparatively less affective [6]. Besides these bacterial strains, there are also some yeast species i.e., Candida krusei and Pichia anomala which were tested and found to have bio-control properties against A. flavus at laboratory level [94]. As these microorganisms having the potential to control AFs under field conditions, so, there is a great need for these trains to be tested under natural environment in the field. One big threat in applying the biological control is due to the presence of biological control agent, where A. flavus and other related AFs producing fungi may accelerate their reproduction and start re-arrangement of their genes, which will enable them to beat the biological control strength. This threat alarm us that providing a biological control strategy, it is very crucial that it should be a complete and a robust one. We must engineer our biological control agent by keeping in mind that there may be re-invasion from the fungus with more threatening capability, due to the rearrangement of its genome overtime. So, the need is to develop more highly sophisticated and defensive strategy for the near future in terms of biological control [40]. Some promising achievements have also been made in term of biological control of AFs contamination by using the nontoxigenic competitive strains of A. parasiticus and A. flavus. Due to this strategy 70 to 90 % reduction in AFs contamination reported in peanut and cotton fields [6, 95, 96]. Owing to these successes two products obtained from nontoxigenic strains have got the approval from US Environmental Protection Agency (EPA) and are being under use as a bio-pesticides in peanut and cotton fields in different states of the US [6].
4.4 Application of advanced agricultural practices
Advanced agricultural practices include timely planting, which can be very helpful to escape the invision of the fungus, maintaning proper plant to plant distances, providing such a conditions in which there is no threat of drought, supply of all essential nutrients at proper time, controlling weed to such a level having no adverse affect on main crop. Similarly, providing strategies which will help to control different insect pest. More importantly, to have proper and on time harvesting. These strategies will greatly minimize the level of AFs contamination at both field and under storage conditions [97, 98]. Crop rotation practices with time to time, proper disposal and management system for the crop residues can also provide assistance in control of A. flavus for upcomimg season crop. Among other nutrients calcium is so much vital for peanut, because it is responsible for the thickness of the peanut cell wall and speeding up the processes of pod filling. Similarly, in case of peanut 50 to 90 % AFs reduction can be obtain through applying lime to the soil, using residues of cereal crops and applying farm yard manure. The farm yard manure helps to provide optimal growth conditions for various beneficial microorganisms having important role in suppressing the soil infections [97, 99, 100]. These strategies are extremely important because they are beneficial and are not so much demanding in terms of cost as well as these are environmental friendly. So, these can be used more frequently as compared to others, to minimize the contamination of various peanut products by AFs.
4.5 The role of classical plant breeding in reducing AFs contamination
An important way to win the combat against A. flavus infection and AFs contamination, is to develop highly resistant cultivars. But unfortunately, resistant cultivar development is a bootleneck in case of peanut due to its narrow genetic diversity. There is also great need of robust, consistent and an efficient techiques for screening the available resources. Various efforts have been made to find indirect methods to select resistant genotypes against pre-harvest contamination by AFs. The aim was to cut down the price, spent on screening various product for removing the contaminated ones [13, 101]. The level of significance of any resistant genotype mainly depends upon that how much it is stable. Resistant genotypes against A. flavus and other related species are very important to control the production of these toxins, but the genetic mechanism underlying this resistance is still thirsty for elucidation. On theoritical scale, important interaction have been found between the resistant genotype and the environment, but at field level they are not so significant. The great obstacle in development of a good resistant backup source against AFs is the that the allelic association among different sources for resistance traits, that can be helpful for breeders to pyramid the non-allelic genes for each resistant mechanism, is still unknown. Under laboratory conditions some promissing results obtained but they were not satisfactory when tested under field conditions [102, 103]. There is a great need to find resistant source which will give stable results under both laboratory and field conditions.
4.6 The role of Biotechnology in combat against AFs contamination
Bringing resistance in genotypes against AFs through classical methods is not so much an efficient and fast to get rid of these toxins. As a ray of hope, biotechnology offer more fast and more efficient way to win the contest against AFs. Through biotechnology, we can get help through studying the three main aspects related to AFs contamination, a): To further strengthen our knowledge about the mechanism of AFs biosynthesis i.e., knowledge of the fungus, b): Knowledge about the environmental factors which are involved in AFs productions and c): How to bring host-plant resistance.
4.6.1 Knowledge about Aspergillus
There is a great need to fully explore and understand each and every aspect of the life cycle of different fungi, responsable for the production of AFs. Currently, the available litrature and research has been extensively reviewed and various future possibilities has been predicted. Valuble progress has been made in exploring biosynthetic pathway of the AFs and several genes have been figure out to have role in AFs production pathway. Several enzymes which speed up this production system and other regulatory systems, have been figured out [71, 104, 105]. Genome editing and manipulation has been carried out to control and guide the AFs production regulation with in the fungus. At more advanced level different genes are identified and cloned having significant role in the biosythesis pathway of the AFs production. These genes could be used to inhibit the biosynthesis pathway of AFs production. Two important Aspergillus species i.e., A. flavus and A. parasiticus have been mapped and sequenced in pinning down a 75 kb gene family, which contain about thirty genes. These genes control the AFs biosynthesis pathway [14, 104, 106]. These information has greatly facilitated and opened the opportunity to find out resistant mechanisms which will prevent fungal progression as well as the production pathway of AFs.
4.6.2 Environmental factors responsible
Environmental factor are of so much importance, greatly affecting AFs production. Among the environmental factors, drought is the major one effects both AFs production as well as appropriate development of seeds [107]. Drought causes reduction in moisture level of the seed as a result of which the property of seed hormone to produce phytoalexins is greatly reduced. Due to the reduction of these phytoalexins, fungal infection occure which cause great economic losses [14]. So, understanding the interaction between the fungus and different environmental factors will be very friutful to minimize the risk posed by AFs contamination.
4.6.3 Mechanism of host-plant resistance
From decades plant breeders and reasearchers are trying to minimize and get rid of AFs contamination through conventional techniques of plant breeding. Although they also have got some promising results but the progress is still very limited to win the combat against Aspergillus and AFs. So, the need to overcome this problem as soon as possible has shifted the research trust from classical breeding to modren plant biotechnology. Through genomic manipulation techniques acompanied by good agricultural practices, provides some golden spark to prevent AFs contamination. In the modern era in various techniques like microarray, sequencing of the whole genome and expressed sequence tags (EST), valuable achivements have gained through finding out different genes involved in host-plant interaction as well as AFs contamination. A few plant factors are also found to have some defensive properties against Aspergillus infection. This defensive nature of the plant is found in three form i.e., Seed protiens which are involved in defense against host cell wall degrading enzymes of the fungus, some natural products found in the seeds or kernels which have a vital role minimize the fungal growth (AFs production) and some other protiens which comes in activation when the plant is under stress conditions [91].
4.6.4 Candidate resistant genes, key towards permanent solution of AFs
Permanent and most effective way to control AFs production is to find out some inhibitory compound against them which may be inhibitory proteins, small molecular weight polypeptides, lectins, hydrolases and cell-surface glycoproteins. Through cDNA several genes are found to have resistance against AFs contamination when they were up-regulated. Similarly, some valuable achievement has been made through proteomic methods [108-110]. In a study carried out by Guo et al. [109] more than 21,777 expressed sequenced tags (ESTs) were generated in peanut to figure out resistant genes, having role in the host plant defense mechanism against Aspergillus and AFs contamination. These genes were then used to develop markers and genetic maps. These genes can be used to profile the transcript and find out candidate genes for various traits of interest. Studies have been carried out to identify the miRNA having role in the gene expression during post-transcriptional stage. Gene expression in resistant peanut cultivar as well as that of susceptible one was profiled and 62 genes were found to have resistance against Aspergillus when they were up-regulated. Along with these 62 genes other twenty-two putative resistant genes against Aspergillus were figured out. In resistant cultivars these genes were highly expressed as compared to that of susceptible one [111].
4.6.5 Targeting Induced local lesions in genomes (TILLING)/mutagenesis
Germplasm are the backup tool for any crop having a good source of resistance to different types of stress conditions and various infectious diseases, having variability among them for the same trait. Sometime the natural variation found among these germplasm is not enough to cope with the sudden invasion from different insect pest and diseases. So, to add to the natural capabilities of the germplasm induced mutation play a very vital role. Various sources have been developed of induced mutation for peanut. Knoll et al. [112] using TILLING techniques screened 3400 mutant lines which were developed through Ethylmethane Sulfonate (EMS) application. This population developed through TILLING will be helpful for functional studies of the genomic, also for recovering unwanted and unintentional mutations [112]. Similarly, using this technique we can also develop such mutated lines which will have the ability to resist against Aspergillus infection and AFs contamination.
4.7 Molecular breeding, a competent approach to get rid of AFs
Classical breeding no doubt has its own importance but molecular breeding has played and still playing an important role in coping with the growing food demand from the rapidly growing population of the world. Molecular markers are the keys of molecular breeding which have enabled the breeders to transfer a trait like resistance to a variety which was susceptible prior to this transformation. These tools can be used to limit the traits which are undesirable. These techniques are so much important to get rid of the Aspergillus and AFs contamination [113]. In case of peanut, very minute variability has been explored for AFs resistance at DNA level using molecular markers, even various agro-morphological traits have also been found with very negligible variation among the various cultivars. At earlier attempts SDS-PAGE was used to find out some differences at protein level to AFs resistance but no promising success was gained. After that an Amplified Fragment Length Polymorphism (AFLP) marker, having resistance to Aspergillus infection, was converted to Sequence Characterized Amplified Region (SCAR) to get promising results in future breeding programs. Then many cultivars of peanut, having promising resistance to AFs, have been used to develop such SCAR markers and one of those having promising results i.e., SCAR “AFs-412” [114]. So, using such type of molecular markers in future breeding programs we can minimize the contamination level of AFs. These markers will be helpful in screening the germplasm prior to the development of commercial peanut cultivars.
The key tool of modern biotechnology and molecular breeding is the ability of altering the genome of an organism which in short terms known as genetic engineering. Through using this technique a molecular plant breeder can bring desirable change in the genetic makeup of a cultivar, which will enable it to survive various stress conditions and show resistance to different invading insect pests. Using molecular transformation tools, we can develop such peanut cultivars which will have resistance to AFs prodeuction [115, 116]. Currently scientists are working on various genes and genes constructs to develop resistant cultivars against those fungi which are responsible for AFs production. Some lytic peptides have been found which are capable of inhibiting A. flavus and are the ray of hope to develop resistant cultivars to AFs contamination. These lytic peptides include D4E1 and D5C, but incase of natural lytic peptide, used to develop stable resistant cultivars, researchers are facing the controversory of effecting non-targeted organism in case of transgene escape to the natural environment [108, 117].
4.7.1 QTL mapping, a golden gift of molecular breeding to mitigate AFs contamination
QTL mapping is one of the golden gifts provided by molecular breeding and is comprehensively under use in modern plant breeding to improve different quality and quantity traits. Most of the important morphological and physilogical traits are controlled by several genes working in groups called quantitative traits. These traits are also called complex, multifactorial or polygenic traits. The genomic regions which control the expression of these complex traits are called QTLs. QTL mapping is one of the reasons which shifted the thinking from classical breeding to molecular breeding and one of the great breakthrough in that respect was the development of molecular or DNA markers. DNA markers are the building blocks of genetic linkage map, while these maps have a critical role to find out the specific genomic regions which control the expression of the quantitive traits [118]. Through QTL analysis tightly linked molecular markers to the trait of interest like A. flavus resistance can be developed and will be deployed in molecular breeding to develop resistant cultivars, and also to screen the susceptible ones. QTL mapping in peanut was slow compared to other crops, because of its complicated genome, but recently due to the advent of more advanced sequencing techniques like Specific Length Amplified Fragment sequencing (SLAF-seq), this process accelerated. SLAF-seq technology is a new, highly precise and robust as compare to other sequencing techniques. More importantly its cost is much lower than its counterparts. It is a combination of locus-specific amplification and high-throughput sequencing, been subjected to a series of critical trials to assure its high accuracy, efficiency, and density. Being in its young age, SLAF-seq has been fruitfully applied to construct high-density genetic map and important QTLs have been identified harboring significant putative candidate genes for different traits of interest in various crops and animal [119-128]. Till now in peanut, very few QTL mapping studies have been reported against AFs contamination. The first study based on QTL mapping against A. flavus invasion reported six QTLs located on chromosome A01, A02, A03, A04, B05, and B08 which contributed 22.7 %, 11.2 %, 6.2 %, 6.6 %, 10.5 %, and 7.3 % in PVE, respectively [129]. Individual QTLs were identified for aflatoxin AFB1, AFB2, and PSII via a RIL population obtained from crossing Zhonghua 10 and ICG 12625. In this study, they identified two QTLs for PSII one located on chromosome A03 sharing 8.0 % in phenotypic variation (PVE) and second located on chromosome A10 with 13.0 % PVE. For AFB1, 7 QTLs were mapped including two major QTLs located on chromosome A05 and B06 sharing 17.9 and 16.3 % in PVE, respectively. For AFB2 they also mapped seven QTLs located on chromosome A07, B05, B06 and B07 with PVE contribution of 12.2 %, 11.1 %, 21.0 % and 14.5 %, respectively [130]. QTL mapping via a RIL population obtained from a cross of two highly contrasting nature to A. flavus resistance Yueyou 92 (YY92) and Xinhuixiaoli (XHXL) during in-vitro seed colonization (IVSC) mapped two major QTLs located on chromosome A03 and B04 shared 19.0 % and 5.1 % in PVE, respectively (unpublished data). Genome-wide association studies using ICRISAT reference set identified a marker associated with IVSC and with more than 24.7 % contribution in PVE [131]. Even these studies are of great importance and the QTLs/ genomic regions identified (Table 1) can be used in future studies to bridge the gap of AFs contamination in peanut but still these findings are very few and further findings needed to find solid and consistent solution to this alarming problem.
Table 1: Mapped QTL of AFs resistance in peanut
|
Trait |
LG |
Position |
Marker Interval |
LOD |
PVE % |
Reference |
|
Resistance to A. flavus invasion |
A01 |
20.35 |
TC11H06–TC4H07 |
4.30 |
22.7 |
Liang et al. 2009 |
|
|
A02 |
9.31 |
gi-716–TC1E05 |
2.26 |
11.2 |
|
|
|
A03 |
5.31 |
pPGSseq18E7–Seq4E08 |
2.60 |
6.2 |
|
|
|
A04 |
12.76 |
pPGPseq2H8–PM3 |
2.1 |
6.6 |
|
|
|
B05 |
25.01 |
pPGPseq7G2–TC5A06 |
2.91 |
10.5 |
|
|
|
B08 |
6.78 |
TC11A04–PM137 |
2.4 |
7.3 |
|
|
Percent seed infection index |
A03 |
28.5 |
AHGS2058- AGGS0052 |
3.1 |
8.0 |
Yu et al. 2019 |
|
|
A10 |
43.5 |
AGGS1425 - ARS710 |
5.0 |
13.0 |
|
|
Aflatoxin B1 |
A05 |
51.1 |
AHGS1245- AGGS0876 |
3.2 |
8.0 |
|
|
|
A05 |
80.3 |
ARS734 - GM2156 |
6.0 |
17.9 |
|
|
|
B06 |
42.5 |
AGGS1515 - AGGS1587 |
6.4 |
16.3 |
|
|
|
B06 |
69.5 |
AHGS1464 - HAS0969 |
3.1 |
7.8 |
|
|
|
B07 |
39.2 |
AGGS1581 - GM2067 |
3.6 |
8.5 |
|
|
|
B07 |
86.0 |
TC3B4 - AHGS2233 |
3.1 |
7.3 |
|
|
|
B07 |
103.7 |
AGGS1081 - AhTE0615 |
3.2 |
7.5 |
|
|
Aflatoxin B2 |
A03 |
50.2 |
AGGS1139 - AHGS2025 |
3.5 |
8.3 |
|
|
|
A07 |
74.3 |
AHGS1454 - HAS1360 |
4.0 |
10.8 |
|
|
|
A07 |
83.5 |
ARS734 - GM2156 |
5.1 |
12.2 |
|
|
|
B05 |
45.4 |
AGGS0979 - TC19E1 |
4.9 |
11.1 |
|
|
|
B06 |
43.1 |
GM2444 - AHGA335472 |
3.8 |
9.3 |
|
|
|
B06 |
43.2 |
GM2444 - AGGS0983 |
8.8 |
21.0 |
|
|
|
B07 |
80.8 |
TC3B4 - AHGS2233 |
5.3 |
14.5 |
|
|
In-Vitro seed colonization |
A03 |
1.673 |
Marker8555604- 8633509 |
10.54 |
19.03 |
(unpublished data) |
|
|
B04 |
1.338 |
Marker4154940- 4158241 |
2.85 |
5.15 |
|
|
In-Vitro seed colonization |
- |
- |
gnPt-737044 (DArT) |
- |
24.7 |
Pandey et al. 2014 |
4.8 Phenotyping, a compulsory call for AFs resistance evaluation
The biosynthesis of AFs in almost all crops is a result of intricate fungus-environment interactions. High level of AFs contamination at field has been reported when the growing season accompanied by drought and hot weather conditions [132]. Studies have shown that maize crop grown under optimum irrigation resulted in reduced fungal infection and AFs contamination, especially when the irrigations were applied in drought stress conditions [133, 134]. Similarly, greater AFs contamination was shown in peanuts during drought-stressed conditions accompanied by high soil temperatures, moreover affecting pre-harvest infection [135]. Mostly it was found that the genotypes reported as resistant under in vitro conditions when tested in natural environment in the field were not so much promising. This threat further compels the need to develop such a high throughput phenotyping assays which will provide field-like environmental conditions for resistance evaluation against AFs [136, 137]. Keeping in mind that nothing is impossible in science, and more importantly due to our lanched sequenced genome for cultivated peanut (http://peanutgr.fafu.edu.cn) [138], we are hopeful that in near future a promising and consistant solution to the problem of AFs contamination may be found.
AFs contamination is an extremely intricate problem which is strongly influenced by numerous external factors as well as genetic resistance. Resistant cultivar development is still being a challenging job in peanut. In addition, to prevent AFs contamination in peanut good management practices during pre-and post-harvest stages are extremely critical. In the past various teachniques have been applied to minimize and eridicate the AFs contamination. Though some encouraging achivements have been made but non of them was found to have 100 % efficacy in elimination of AFs contamination, where the main hurdle found was the availability of very little knowledge of the molecular mechanism of AFs production. As alternative various conventional breeding techniques and stratigies were found helfpul up to some extent but no one was found adequate for complete solution of this problem. To further speed up the contest against AFs contamination a sound and deep investigation is required at molecular level to find out more resistant genes againt AFs production. Significant control of AFs contamination needs a multipronged method comprise of biological control, more advance agronomic and cultural practices along with high genetic resistance by the host plant. Following are some simple recommendations which can reduce AFs contamination to significant level: Use of lime (0.5 t/ha), cereal crop residue (5 t/ha) and farm yard manure (10t/ha) at sowing time to reduce A. flavus infection and AFs contamination from 50 to 90 % [139]. Select comparatively highly resistant cultivar to A. flavus infection and AFs contamination and with better tolerance to drought [140]. Harvest the crop at optimum maturity. Harvesting before optimum maturity or making delay in harvesting causing poor quality seed which provide opportunity for A. flavus to produce AFs. Try to avoid any physical damage to pods at the time of harvest due to which A. flavus infection occurs more frequently. For drying the harvested pods, clean sheets should be used instead of direct drying on the ground. Better to remove the immature and infected pods before drying from the mature and healthy pods. Sustain proper storage facilities having proper ventilation. The pods should be dried and more importantly have low relative humidity. Having proper control on the entry exit of insect pest and rodents.
Authors are thankful to Fujian Key Laboratory of Plant Molecular and Cell Biology, Fujian Agriculture and Forestry University, Fuzhou, Fujian, 350002, China, for providing research facilities and technical guidance.
AUTHOR CONTRIBUTION STATEMENT
SAK wrote the manuscript and ZW modified the manuscript.
Pandey, M.K., E. Monyo, P. Ozias-Akins, et al., Advances in Arachis genomics for peanut improvement. Biotechnology Advances, 2012. 30(3): p. 639-651. PMid:22094114
View Article PubMed/NCBIVarshney, R.K., S.M. Mohan, P.M. Gaur, et al., Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnology advances, 2013. 31(8): p. 1120-1134. PMid:23313999
View Article PubMed/NCBIJambunathan, R., S. Hall, P. Sudhir, et al., Uses of Tropical Grain Legumes Proceedings of a Consultants Meeting 27-30 Mar 1989. 1991.
Payne, G.A. and N.W. Widstrom, Aflatoxin in maize. Critical Reviews in Plant Sciences, 1992. 10(5): p. 423-440.
View ArticleCastegnaro, M. and D. McGregor, Carcinogenic risk assessment of mycotoxins. Revue de Medecine Veterinaire, 1998.
Dorner, J.W., Biological control of aflatoxin contamination of crops. Journal of Toxicology: Toxin Reviews, 2004. 23(2-3): p. 425-450.
View ArticleVan Egmond, H.P. and M.A. Jonker, Worldwide regulations on aflatoxins-the situation in 2002. Journal of Toxicology: Toxin Reviews, 2004. 23(2-3): p. 273-293.
View ArticleLiang, X., M. Luo, and B. Guo, Resistance Mechanisms to Aspergillus flavus Infection and Aflatoxin Contamination in Peanut (Arachis hypogaea). Plant Pathology Journal, 2006. 5(1): p. 115-124.
View ArticlePassone, M.A., M. Ruffino, V. Ponzio, et al., Postharvest control of peanut Aspergillus section Flavi populations by a formulation of food-grade antioxidants. International journal of food microbiology, 2009. 131(2-3): p. 211-217. PMid:19339073
View Article PubMed/NCBISmith, J.E., G. Solomons, C. Lewis, et al., Role of mycotoxins in human and animal nutrition and health. Natural Toxins, 1995. 3(4): p. 187-192. PMid:7582615
View Article PubMed/NCBIGuo, B., C. Chen, Y. Chu, et al., Advances in genetics and genomics for sustainable peanut production. Sustainable Agriculture New Biotechnologies, 2011: p. 341-367.
View ArticleGuo, B., J. Yu, C.C. Holbrook, et al., Strategies in prevention of preharvest aflatoxin contamination in peanuts: aflatoxin biosynthesis, genetics and genomics. Peanut Science, 2009. 36(1): p. 11-20.
View ArticleHolbrook, C., C. Kvien, K. Rucker, et al., Preharvest aflatoxin contamination in drought-tolerant and drought-intolerant peanut genotypes. Peanut Science, 2000. 27(2): p. 45-48.
View ArticleGuo, B., C. Holbrook, J. Yu, et al., Application of technology of gene expression in response to drought stress and elimination of preharvest aflatoxin contamination. Aflatoxin Food Safety, 2005. 26: p. 313-331.
View ArticleCraufurd, P., P. Prasad, F. Waliyar, et al., Drought, pod yield, pre-harvest Aspergillus infection and aflatoxin contamination on peanut in Niger. Field Crops Research, 2006. 98(1): p. 20-29.
View ArticleWilliams, J.H., T.D. Phillips, P.E. Jolly, et al., Human aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. The American journal of clinical nutrition, 2004. 80(5): p. 1106-1122. PMid:15531656
View Article PubMed/NCBIWilliams, J.H., J.A. Grubb, J.W. Davis, et al., HIV and hepatocellular and esophageal carcinomas related to consumption of mycotoxin-prone foods in sub-Saharan Africa. The American journal of clinical nutrition, 2010. 92(1): p. 154-160. PMid:20484447
View Article PubMed/NCBIWogan, G.N., Chemical nature and biological effects of the aflatoxins. Bacteriological reviews, 1966. 30(2): p. 460.
Azziz-Baumgartner, E., K. Lindblade, K. Gieseker, et al., Case-Control Study of an Acute Aflatoxicosis Outbreak, Kenya, 2004. Environmental health perspectives, 2005. 113(12): p. 1779-1783. PMid:16330363
View Article PubMed/NCBIDiener, U.L., R.J. Cole, T. Sanders, et al., Epidemiology of aflatoxin formation by Aspergillus flavus. Annual Review of Phytopathology, 1987. 25(1): p. 249-270.
View ArticleEaton, D.L. and J.D. Groopman, The toxicology of aflatoxins: human health, veterinary, and agricultural significance. 2013: Elsevier.
Reddy, K., B. Salleh, B. Saad, et al., An overview of mycotoxin contamination in foods and its implications for human health. Toxin reviews, 2010. 29(1): p. 3-26.
View ArticleCotty, P., Virulence and cultural characteristics of two Aspergillus flavus strains pathogenic on cotton. Phytopathology, 1989. 79(7): p. 808-814.
View ArticleNozawa, K., S. Sekita, M. Harada, et al., Isolation and Structures of Two New Indoloditerpenes Related to Aflavinine from a Microsclerotium-Producing Strain of Aspergillus flavus. Chemical Pharmaceutical Bulletin, 1989. 37(3): p. 626-630.
View ArticleSaito, M. and O. TSURUTA, A new variety of Aspergillus flavus from tropical soil in Thailand and its aflatoxin productivity. JSM Mycotoxins, 1993. 1993(37): p. 31-36.
View ArticleGeiser, D.M., J.W. Dorner, B.W. Horn, et al., The phylogenetics of mycotoxin and sclerotium production in Aspergillus flavus and Aspergillus oryzae. Fungal Genetics Biology, 2000. 31(3): p. 169-179. PMid:11273679
View Article PubMed/NCBIFrisvad, J.C., P. Skouboe, and R.A. Samson, Taxonomic comparison of three different groups of aflatoxin producers and a new efficient producer of aflatoxin B1, sterigmatocystin and 3-O-methylsterigmatocystin, Aspergillus rambellii sp. nov. Systematic applied microbiology, 2005. 28(5): p. 442-453. PMid:16094871
View Article PubMed/NCBIEgel, D.S., P.J. Cotty, and K.S. Elias, Relationships among isolates of Aspergillus sect. Flavi that vary in aflatoxin production. pathology, 1994. 84906: p. 912.
View ArticleGibson, A.M., J. Baranyi, J.I. Pitt, et al., Predicting fungal growth: the effect of water activity on Aspergillus flavus and related species. International journal of food microbiology, 1994. 23(3-4): p. 419-431. 90167-8
View ArticleVujanovic, V., W. Smoragiewicz, and K. Krzysztyniak, Airborne fungal ecological niche determination as one of the possibilities for indirect mycotoxin risk assessment in indoor air. Environmental Toxicology: An International Journal, 2001. 16(1): p. 1-8. 16:1<1::AID-TOX10>3.0.CO;2-8
View ArticlePayne, G. and M. Brown, Genetics and physiology of aflatoxin biosynthesis. Annual review of phytopathology, 1998. 36(1): p. 329-362. PMid:15012504
View Article PubMed/NCBIYu, J. and D. Arora, Genetics and biochemistry of mycotoxin synthesis. Fungal biotechnology in agricultural, food, environmental applications, 2004. 21: p. 343-361.
View ArticleCuero, R., T. Ouellet, J. Yu, et al., Metal ion enhancement of fungal growth, gene expression and aflatoxin synthesis in Aspergillus flavus: RT‐PCR characterization. Journal of applied microbiology, 2003. 94(6): p. 953-961. PMid:12752802
View Article PubMed/NCBIMaggio-Hall, L.A., R.A. Wilson, and N.P. Keller, Fundamental contribution of β-oxidation to polyketide mycotoxin production in planta. Molecular plant-microbe interactions, 2005. 18(8): p. 783-793. PMid:16134890
View Article PubMed/NCBIBrodhagen, M. and N.P. Keller, Signalling pathways connecting mycotoxin production and sporulation. Molecular Plant Pathology, 2006. 7(4): p. 285-301. PMid:20507448
View Article PubMed/NCBIKim, J.H., B.C. Campbell, J. Yu, et al., Examination of fungal stress response genes using Saccharomyces cerevisiae as a model system: targeting genes affecting aflatoxin biosynthesis by Aspergillus flavus Link. Applied Microbiology Biotechnology, 2005. 67(6): p. 807-815. PMid:15614562
View Article PubMed/NCBIKim, J.H., B. Campbell, R. Molyneux, et al., Gene targets for fungal and mycotoxin control. Mycotoxin research, 2006. 22(1): p. 3-8. PMid:23605494
View Article PubMed/NCBIHicks, J.K., J.H. Yu, N.P. Keller, et al., Aspergillus sporulation and mycotoxin production both require inactivation of the FadA Gα protein‐dependent signaling pathway. The EMBO Journal, 1997. 16(16): p. 4916-4923. PMid:9305634
View Article PubMed/NCBIChang, P.-K., C.D. Skory, and J.E. Linz, Cloning of a gene associated with aflatoxin B1 biosynthesis in Aspergillus parasiticus. Current genetics, 1992. 21(3): p. 231-233. PMid:1563048
View Article PubMed/NCBIEhrlich, K.C., Non-aflatoxigenic Aspergillus flavus to prevent aflatoxin contamination in crops: advantages and limitations. Frontiers in microbiology, 2014. 5: p. 50.
View ArticleMahoney, N. and R.J. Molyneux, Phytochemical inhibition of aflatoxigenicity in Aspergillus flavus by constituents of walnut (Juglans regia). Journal of Agricultural Food Chemistry, 2004. 52(7): p. 1882-1889. PMid:15053524
View Article PubMed/NCBIYin, Y.-n., L.-y. Yan, J.-h. Jiang, et al., Biological control of aflatoxin contamination of crops. Journal of Zhejiang University Science B, 2008. 9(10): p. 787-792. PMid:18837105
View Article PubMed/NCBIMagan, N., V. Sanchis, and D. Aldred, Role of spoilage fungi in seed deterioration. Fungal biotechnology in agricultural, food environmental applications, 2004: p. 311-323.
View ArticlePapa, K., Genetics of Aspergillus flavus: linkage of aflatoxin mutants. Canadian journal of microbiology, 1984. 30(1): p. 68-73. PMid:6424919
View Article PubMed/NCBIPapa, K., Genetics of Aspergillus flavus: complementation and mapping of aflatoxin mutants. Genetics Research, 1979. 34(1): p. 1-9. PMid:116906
View Article PubMed/NCBIBennett, J., Microbiological aspects of the Aflatoxin problem. Amer Ass Feed Microscop Offic Proc, 1970.
Bennett, J.W. and L. Goldblatt, The isolation of mutants of Aspergillus flavus and A. parasiticus with altered aflatoxin producing ability. Sabouraudia, 1973. 11(3): p. 235-241. PMid:4203153
View Article PubMed/NCBIBennett, J., L. Lee, and A. Cucullu, Effect of dichlorvos on Aflatoxin and versicolorin A production in Aspergillus parasiticus. Botanical Gazette, 1976. 137(4): p. 318-324.
View ArticleBennett, J., Aflatoxins and anthraquinones from diploids of Aspergillus parasiticus. Microbiology, 1979. 113(1): p. 127-136. PMid:501330
View Article PubMed/NCBIBennett, J., F. Kronberg, L. Goodman, et al., Isolation of an anthraquinone-accumulating mutant of Aspergillus parasiticus and partial characterization by dry column chromatography. Mycologia, 1983: p. 202-208.
View ArticleBennett, J.W., L.S. Lee, and C. Vinnett, The correlation of aflatoxin and norsolorinic acid production. Journal of the American Oil Chemists' Society, 1971. 48(7): p. 368.
View ArticleBennett, J., F. Kronberg, and G. Gougis, Pigmented isolates from anthraquinone-producing mutants of Aspergillus parasiticus. Am. Soc. Microbiol, 1976. 76(6).
Bennett, I., One gene to whole pathway: the Role of. Adv Appl Microbiol, 1997. 45(1). 70260-0
View ArticleHsieh, D.P., M.T. Lin, R.C. Yao, et al., Biosynthesis of aflatoxin. Conversion of norsolorinic acid and other hypothetical intermediates into aflatoxin B1. Journal of agricultural food chemistry, 1976. 24(6): p. 1170-1174. PMid:1002896
View Article PubMed/NCBIDutton, M., Enzymes and aflatoxin biosynthesis. Microbiological reviews, 1988. 52(2): p. 274.
Hsieh, D. and R. Mateles, The relative contribution of acetate and glucose to aflatoxin biosynthesis. Biochimica et biophysica acta, 1970. 208: p. 482-486. 90222-9
View ArticleHsieh, D., M. Lin, and R. Yao, Conversion of sterigmatocystin to aflatoxin B1 by Aspergillus parasiticus. Biochemical biophysical research communications, 1973. 52(3): p. 992-997. 91035-8
View ArticleYu, J., P.-K. Chang, J.W. Cary, et al., Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Applied Environmental Microbiology, 1995. 61(6): p. 2365-2371.
Bennett, J. and K. Papa, The Aflatoxigenic Aspergillus spp. Advances in plant pathology: genetics of plant pathogenic fungi, 1988. 6: p. 263-80.
View ArticleBennett, J., R. Silverstein, and S. Kruger, Isolation and characterization of two nonaflatoxigenic classes of morphological variants of Aspergillus parasiticus. Journal of the American Oil Chemists' Society, 1981. 58(12): p. A952-A955.
View ArticleBhatnagar DE, Ehrlich KC, and C. TE., Oxidation-reduction reactions in biosynthesis of secondary metabolites. Handbook of applied mycology, 1991. 5: p. 255-286.
Chang, P.-K., Lack of interaction between AFLR and AFLJ contributes to nonaflatoxigenicity of Aspergillus sojae. Journal of biotechnology, 2004. 107(3): p. 245-253. PMid:14736460
View Article PubMed/NCBIChang, P.-K., J. Cary, D. Bhatnagar, et al., Cloning of the Aspergillus parasiticus apa-2 gene associated with the regulation of aflatoxin biosynthesis. Applied Environmental Microbiology, 1993. 59(10): p. 3273-3279.
Chang, P.-K., J.W. Cary, J. Yu, et al., The Aspergillus parasiticus polyketide synthase gene pksA, a homolog of Aspergillus nidulans wA, is required for aflatoxin B 1 biosynthesis. Molecular General Genetics, 1995. 248(3): p. 270-277. PMid:7565588
View Article PubMed/NCBIChang, P.-K., J. Yu, D. Bhatnagar, et al., The Carboxy-Terminal Portion of the Aflatoxin Pathway Regulatory Protein AFLR of Aspergillus parasiticus ActivatesGAL1:: lacZ Gene Expression in Saccharomyces cerevisiae. Applied environmental microbiology, 1999. 65(6): p. 2508-2512.
Cleveland, T.E., A. Lax, L. Lee, et al., Appearance of enzyme activities catalyzing conversion of sterigmatocystin to aflatoxin B1 in late-growth-phase Aspergillus parasiticus cultures. Applied environmental microbiology, 1987. 53(7): p. 1711-1713.
Crawford, J.M., P.M. Thomas, J.R. Scheerer, et al., Deconstruction of iterative multidomain polyketide synthase function. Science, 2008. 320(5873): p. 243-246. PMid:18403714
View Article PubMed/NCBIEhrlich, K.C., J.W. Cary, and B.G. Montalbano, Characterization of the promoter for the gene encoding the aflatoxin biosynthetic pathway regulatory protein AFLR. Biochimica et Biophysica Acta -Gene Structure, 1999. 1444(3): p. 412-417. 00022-6
View ArticleKeller, N., H. Dischinger, D. Bhatnagar, et al., Purification of a 40-kilodalton methyltransferase active in the aflatoxin biosynthetic pathway. Applied environmental microbiology, 1993. 59(2): p. 479-484.
Ehrlich, K.C. and J. Yu, Aflatoxin-like gene clusters and how they evolved, in Mycotoxins in food, feed and bioweapons. 2009, Springer. p. 65-75.
View ArticleEhrlich, K.C., Predicted roles of the uncharacterized clustered genes in aflatoxin biosynthesis. Toxins, 2009. 1(1): p. 37-58. PMid:22069531
View Article PubMed/NCBIYu, J., P.-K. Chang, K.C. Ehrlich, et al., Clustered pathway genes in aflatoxin biosynthesis. Applied environmental microbiology, 2004. 70(3): p. 1253-1262. PMid:15006741
View Article PubMed/NCBIChang, P.-K., B.W. Horn, and J.W. Dorner, Sequence breakpoints in the aflatoxin biosynthesis gene cluster and flanking regions in nonaflatoxigenic Aspergillus flavus isolates. Fungal Genetics Biology, 2005. 42(11): p. 914-923. PMid:16154781
View Article PubMed/NCBIWilson, D.M., Analytical methods for aflatoxins in corn and peanuts. Archives of environmental contamination, 1989. 18(3): p. 308-314. PMid:2730149
View Article PubMed/NCBITrail, F., N. Mahanti, and J.J.M. Linz, Molecular biology of aflatoxin biosynthesis. 1995. 141(4): p. 755-765. PMid:7773383
View Article PubMed/NCBITrail, F., N. Mahanti, M. Rarick, et al., Physical and transcriptional map of an aflatoxin gene cluster in Aspergillus parasiticus and functional disruption of a gene involved early in the aflatoxin pathway. Applied Environmental Microbiology, 1995. 61(7): p. 2665-2673.
Townsend, C., Progress toward a biosynthetic rationale of the aflatoxin pathway. Pure Applied Chemistry, 1986. 58(2): p. 227-238.
View ArticleYu, J., D. Bhatnagar, and T.E. Cleveland, Completed sequence of aflatoxin pathway gene cluster in Aspergillus parasiticus. Febs Letters, 2004. 564(1-2): p. 126-130. 00327-8
View ArticleWarburton, M.L. and W.P. Williams, Aflatoxin resistance in maize: what have we learned lately? Advances in Botany, 2014.
View ArticleUpadhyaya, H., S. Nigam, and R. Thakur, Genetic enhancement for resistance to aflatoxin contamination in groundnut. 2002.
Garrido-Bazan, V., G. Mahuku, M. Bibbins-Martinez, et al., Dissection of mechanisms of resistance to Aspergillus flavus and aflatoxin using tropical maize germplasm. World Mycotoxin Journal, 2018. 11(2): p. 215-224.
View ArticleLaPrade, J., J. Bartz, A. Norden, et al., Correlation of peanut seed-coat surface wax accumulations with tolerance to colonization by Aspergillus flavus. J Amer Peanut Res Educ Soc, 1973. 5: p. 89-94.
Liang, X., G. Zhou, and R. Pan, Study on the relationship of wax and cutin layers in peanut seeds and resistance to invasion and aflatoxin production by Aspergillus flavus. J Tropic Subtropic Bot, 2003. 11: p. 11-14.
Liang, X., M. Luo, and B. Guo, Resistance Mechanisms to aspergillus falvus infectin and aflatoxin contaminatin in peanut (Arachis hypogaea). Plant Pathology Journal, 2006. 5(1): p. 115-124.
View ArticleLiang, X., Studies on the mechanism and inheritance of resistance to Aspergillus flavus invasion and aflatoxin production in peanut (Arachis hypogaea L.). 2002, Ph. D. Thesis, University of South China University, Guangzhou, China.
Choudhary, A.K. and P. Kumari, Management of mycotoxin contamination in preharvest and post harvest crops: present status and future prospects. Journal of Phytology, 2010.
Bhatnagar, D., P. Cotty, and T. Cleveland, Genetic and biological control of aflatoxigenic fungi. Microbial food contamination, 2001: p. 208-240.
Cleveland, T., J. Yu, Z.-y. Chen, et al. The use of crop proteomics and fungal genomics in elucidating fungus-crop interactions. in Proceedings of the Myco-Globe Conference. 2006.
Girdthai, T., S. Jogloy, N. Vorasoot, et al., Associations between physiological traits for drought tolerance and aflatoxin contamination in peanut genotypes under terminal drought. Plant Breeding, 2010. 129(6): p. 693-699.
View ArticleAnderson, W., C. Holbrook, D. Wilson, et al., Evaluation of preharvest Aflatoxin contamination in several potentially resistant peanut genotypes. Peanut Science, 1995. 22(1): p. 29-32.
View ArticleMehan, V. Screening groundnuts for resistance to seed invasion by Aspergillus flavus and to aflatoxin production [a review]. in International Workshop on Aflatoxin Contamination of Groundnut, Patancheru, AP (India), 6-9 Oct 1987. 1989. ICRISAT.
Palumbo, J.D., J.L. Baker, and N.E. Mahoney, Isolation of bacterial antagonists of Aspergillus flavus from almonds. Microbial ecology, 2006. 52(1): p. 45-52. PMid:16767519
View Article PubMed/NCBINesci, A.V., R.V. Bluma, and M.G. Etcheverry, In vitro selection of maize rhizobacteria to study potential biological control of Aspergillus section Flavi and aflatoxin production. European journal of plant pathology, 2005. 113(2): p. 159-171.
View ArticleMasoud, W. and C.H. Kaltoft, The effects of yeasts involved in the fermentation of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. International journal of food microbiology, 2006. 106(2): p. 229-234. PMid:16213049
View Article PubMed/NCBIPitt, J. and A.D. Hocking, Mycotoxins in Australia: biocontrol of aflatoxin in peanuts. Mycopathologia, 2006. 162(3): p. 233-243. PMid:16944290
View Article PubMed/NCBIDorner, J., Management and prevention of mycotoxins in peanuts. Food Additives Contaminants, 2008. 25(2): p. 203-208. PMid:18286410
View Article PubMed/NCBIWaliyar, F., M. Osiru, H. Sudini, et al., Reducing aflatoxins in groundnuts through integrated management and biocontrol. 2013.
Bruns, H.A., Controlling aflatoxin and fumonisin in maize by crop management. Journal of Toxicology: Toxin Reviews, 2003. 22(2-3): p. 153-173.
View ArticleWaliyar, F., P.L. Kumar, A. Traoré, et al., Pre-and postharvest management of aflatoxin contamination in peanuts. Mycotoxins: detection methods, management, public health agricultural trade. CABI, Wallingford, UK, 2008: p. 209-218.
View ArticleHamidou, F., A. Rathore, F. Waliyar, et al., Although drought intensity increases aflatoxin contamination, drought tolerance does not lead to less aflatoxin contamination. Field Crops Research, 2014. 156: p. 103-110.
View ArticleWindham, G. and W. Williams, Aspergillus flavus infection and aflatoxin accumulation in resistant and susceptible maize hybrids. Plant Disease, 1998. 82(3): p. 281-284. PMid:30856857
View Article PubMed/NCBIWaliyar, F., A. Traore, D. Fatondji, et al., Effect of irrigation interval, planting date, and cultivar on Aspergillus flavus and aflatoxin contamination of peanut in a sandy soil of Niger. Peanut science, 2003. 30(2): p. 79-84.
View ArticleKisyombe, C.T., M. Beute, and G. Payne, Field evaluation of peanut genotypes for resistance to infection by Aspergillus parasiticus. Peanut Science, 1985. 12(1): p. 12-17.
View ArticleYu, J., Current understanding on aflatoxin biosynthesis and future perspective in reducing aflatoxin contamination. Toxins, 2012. 4(11): p. 1024-1057. PMid:23202305
View Article PubMed/NCBIBhatnagar, D., K. Ehrlich, and T. Cleveland, Molecular genetic analysis and regulation of aflatoxin biosynthesis. Applied Microbiology and Biotechnology, 2003. 61(2): p. 83-93. PMid:12655449
View Article PubMed/NCBIEhrlich, K., J. Yu, and P. Cotty, Aflatoxin biosynthesis gene clusters and flanking regions. Journal of Applied Microbiology, 2005. 99(3): p. 518-527. PMid:16108793
View Article PubMed/NCBIKambiranda, D.M., H.K. Vasanthaiah, R.K.A. Ananga, et al., Impact of drought stress on peanut (Arachis hypogaea L.) productivity and food safety, in Plants and environment. 2011, Intech.
Cary, J.W., K. Rajasekaran, R.L. Brown, et al., Developing resistance to aflatoxin in maize and cottonseed. Toxins, 2011. 3(6): p. 678-696. PMid:22069734
View Article PubMed/NCBIGuo, B., X. Chen, P. Dang, et al., Peanut gene expression profiling in developing seeds at different reproduction stages during Aspergillus parasiticus infection. BMC Developmental Biology, 2008. 8(1): p. 12. PMid:18248674
View Article PubMed/NCBIWang, T., E. Zhang, X. Chen, et al., Identification of seed proteins associated with resistance to pre-harvested aflatoxin contamination in peanut (Arachis hypogaea L). BMC Plant Biology, 2010. 10(1): p. 267. PMid:21118527
View Article PubMed/NCBIGuo, B., N.D. Fedorova, X. Chen, et al., Gene expression profiling and identification of resistance genes to Aspergillus flavus infection in peanut through EST and microarray strategies. Toxins, 2011. 3(7): p. 737-753. PMid:22069737
View Article PubMed/NCBIKnoll, J.E., M.L. Ramos, Y. Zeng, et al., TILLING for allergen reduction and improvement of quality traits in peanut (Arachis hypogaea L.). BMC plant biology, 2011. 11(1): p. 81. PMid:21569438
View Article PubMed/NCBIBrown, R.L., A. Menkir, Z.-Y. Chen, et al., Breeding aflatoxin-resistant maize lines using recent advances in technologies-a review. Food Additives Contaminants: Part A, 2013. 30(8): p. 1382-1391. PMid:23859902
View Article PubMed/NCBILei, Y., B.-S. Liao, S.-Y. Wang, et al., A SCAR marker for resistance to Aspergillus flavus in peanut (Arachis hypogaea L.). Yi chuan= Hereditas, 2006. 28(9): p. 1107-1111. PMid:16963420
View Article PubMed/NCBIAmalu, U.C., Plant biotechnology and food crop development in Sub-Saharan Africa. Technology in Society, 2004. 26(4): p. 537-550. 00097-6
View ArticleWani, S.H., Inducing fungus-resistance into plants through biotechnology. Notulae Scientia Biologicae, 2010. 2(2): p. 14-21.
View ArticleMarcos, J.F., A. Muñoz, E. Pérez-Payá, et al., Identification and rational design of novel antimicrobial peptides for plant protection. Annu. Rev. Phytopathol., 2008. 46: p. 273-301. PMid:18439131
View Article PubMed/NCBICollard, B., M. Jahufer, J. Brouwer, et al., An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica, 2005. 142(1-2): p. 169-196.
View ArticleSun, X., D. Liu, X. Zhang, et al., SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PloS one, 2013. 8(3): p. e58700. PMid:23527008
View Article PubMed/NCBIZhang, Y., L. Wang, H. Xin, et al., Construction of a high-density genetic map for sesame based on large scale marker development by specific length amplified fragment (SLAF) sequencing. BMC plant biology, 2013. 13(1): p. 141. PMid:24060091
View Article PubMed/NCBIHuang, S., J. Ding, D. Deng, et al., Draft genome of the kiwifruit Actinidia chinensis. Nature communications, 2013. 4: p. 2640.
Qi, Z., L. Huang, R. Zhu, et al., A high-density genetic map for soybean based on specific length amplified fragment sequencing. PloS one, 2014. 9(8): p. e104871. PMid:25118194
View Article PubMed/NCBIZhang, J., Q. Zhang, T. Cheng, et al., High-density genetic map construction and identification of a locus controlling weeping trait in an ornamental woody plant (Prunus mume Sieb. et Zucc). DNA research, 2015. 22(3): p. 183-191. PMid:25776277
View Article PubMed/NCBIWei, Q., Y. Wang, X. Qin, et al., An SNP-based saturated genetic map and QTL analysis of fruit-related traits in cucumber using specific-length amplified fragment (SLAF) sequencing. BMC genomics, 2014. 15(1): p. 1158. PMid:25534138
View Article PubMed/NCBIXu, X., R. Xu, B. Zhu, et al., A high-density genetic map of cucumber derived from Specific Length Amplified Fragment sequencing (SLAF-seq). Frontiers in plant science, 2015. 5: p. 768.
View ArticleZhu, W.Y., L. Huang, L. Chen, et al., A high-density genetic linkage map for cucumber (Cucumis sativus L.): based on specific length amplified fragment (SLAF) sequencing and QTL analysis of fruit traits in cucumber. Frontiers in plant science, 2016. 7: p. 437.
View ArticleZhao, X., L. Huang, X. Zhang, et al., Construction of high-density genetic linkage map and identification of flowering-time QTLs in orchardgrass using SSRs and SLAF-seq. Scientific reports, 2016. 6: p. 29345. PMid:27389619
View Article PubMed/NCBILiu, C., B. Fan, Z. Cao, et al., Development of a high-density genetic linkage map and identification of flowering time QTLs in adzuki bean (Vigna angularis). Scientific reports, 2016. 6: p. 39523. PMid:28008173
View Article PubMed/NCBILiang, X., G. Zhou, Y. Hong, et al., Overview of research progress on peanut (Arachis hypogaea L.) host resistance to aflatoxin contamination and genomics at the Guangdong Academy of Agricultural Sciences. Peanut science, 2009. 36(1): p. 29-34.
View ArticleYu, B., D. Huai, L. Huang, et al., Identification of genomic regions and diagnostic markers for resistance to aflatoxin contamination in peanut (Arachis hypogaea L.). BMC genetics, 2019. 20(1): p. 32. PMid:30866805
View Article PubMed/NCBIPandey, M.K., H.D. Upadhyaya, A. Rathore, et al., Genomewide association studies for 50 agronomic traits in peanut using the 'reference set'comprising 300 genotypes from 48 countries of the semi-arid tropics of the world. PLoS one, 2014. 9(8): p. e105228.
View ArticlePayne, G. Characterization of inhibitors from corn seeds and the use of a new reporter construct to select corn genotypes resistant to aflatoxin accumulation. in Proc USDA-ARS Aflatoxin Elimination Workshop. 1997.
Guo, B., Z.Y. Chen, R.D. Lee, et al., Drought stress and preharvest aflatoxin contamination in agricultural commodity: genetics, genomics and proteomics. Journal of Integrative Plant Biology, 2008. 50(10): p. 1281-1291. PMid:19017115
View Article PubMed/NCBIKebede, H., H.K. Abbas, D.K. Fisher, et al., Relationship between aflatoxin contamination and physiological responses of corn plants under drought and heat stress. Toxins, 2012. 4(11): p. 1385-1403. PMid:23202322
View Article PubMed/NCBIWaliyar, F., S. Reddy, and R. Thakur, Effects of mycotoxins on cereals grain feed and fodder quality. Alternative Uses of Sorghum Pearl Millet in Asia, 2003. 128.
Davidson Jr, J., R. Hill, R. Cole, et al., Field performance of two peanut cultivars relative to aflatoxin contamination. Peanut Science, 1983. 10(1): p. 43-47.
View ArticleBlankenship, P., R. Cole, and T. Sanders, Comparative susceptibility of four experimental peanut lines and the cultivar Florunner to preharvest aflatoxin contamination. Peanut Science, 1985. 12(2): p. 70-72.
View ArticleZhuang, W., H. Chen, M. Yang, et al., The genome of cultivated peanut provides insight into legume karyotypes, polyploid evolution and crop domestication. Nature genetics, 2019: p. 1.
Karthikeyan, A., Effect of organic amendments, antagonist Trichoderma viride and fungicides on seed and collar rot of groundnut. Plant Disease Research, 1996. 11: p. 72-74.
Okello, D., A. Kaaya, J. Bisikwa, et al., Management of aflatoxins in groundnuts: A manual for farmers, processors, traders and consumers in Uganda. National Agricultural Research Organization, Entebbe, Uganda, 2010.
